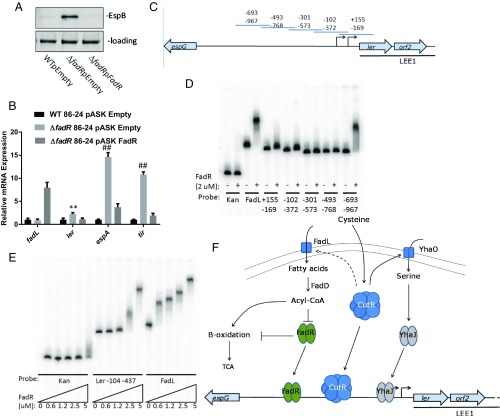
Fig. 4.
FadR LEE regulator in EHEC. (A) Western for EspB from supernatants of WT, ΔfadR, and complemented strains (cultures grown in DMEM). (B) qRT-PCR of LEE genes from WT, ΔfadR, and complemented strains (cultures grown in DMEM). (C) Diagram of EHEC LEE1 promoter segments used for EMSA probes. Probes are designed to have ∼80 bp of overlap between segments, extending from 967 bp upstream of the proximal LEE1 transcription start site to 155 bp downstream. (D) EMSA of His-tagged FadR and kanamycin probe (negative control), fadL promoter probe (positive control), and segments of the EHEC LEE1 regulatory region numbered from the proximal transcriptional start site. (E) EMSA of a Citrobacter LEE1 promoter fragment extending from 437 bp upstream to 104 bp upstream of the transcriptional start site. (F) Schematic of model of fadR- and cutR-dependent LEE regulation. **P < 0.01, ##P < 0.0001.