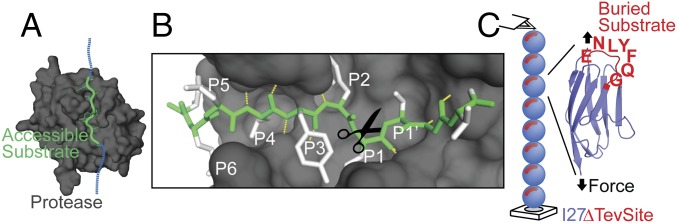
Fig. 1.
Experimental framework. (A) Structure of the Tev protease (gray surface) bound to its substrate (green tube). The additional blue tube represents the contiguous extended chain. For clarity, residues F172 and K2201 are not shown on the surface. (B) P6–P1′ (green) accommodates in the binding-site cleft with a curved backbone geometry stabilized by hydrogen bonds (yellow dashes) orienting the side chains (white) toward the specific pocket. (C) With an atomic force microscope, we applied a stretching force (black arrows) to a single octamer of the Ig domain I27ΔTevSite. Each I27 folded domain (blue) contains a Tev cleavage site ENLYFQG (red). The structure of I27 (PDB ID code 1TIT) shows that the last residue of ENLYFQG corresponds to the native glycine G32 and participates in the β-strand C arrow colored in blue and red. It suggests that the Tev site is not accessible in the folded I27ΔTevSite domain.