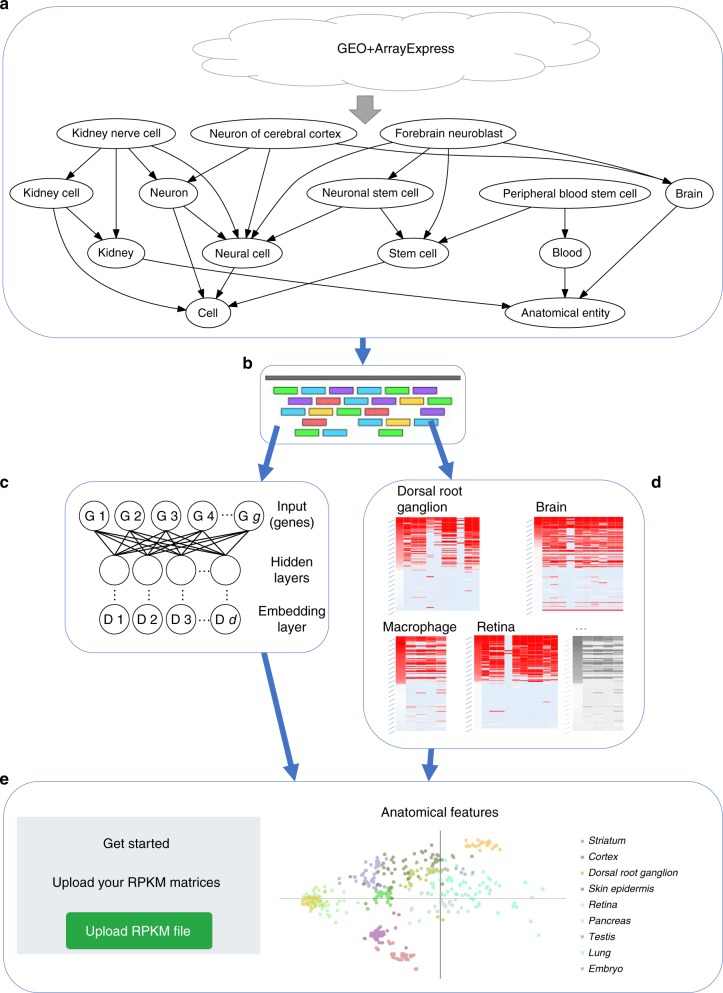
Fig. 1.
Pipeline for large-scale, automated analysis of scRNA-seq data. a Bi-weekly querying of GEO and ArrayExpress to download the latest data, followed by automatic label inference by mapping to the Cell Ontology. b Uniform alignment of all datasets using HISAT2, followed by quantification to obtain RPKM values. c Supervised dimensionality reduction using our neural embedding models. d Identification of cell-type-specific gene lists using differential expression analysis. e Integration of data and methods into a publicly available web application