Fig. 1.
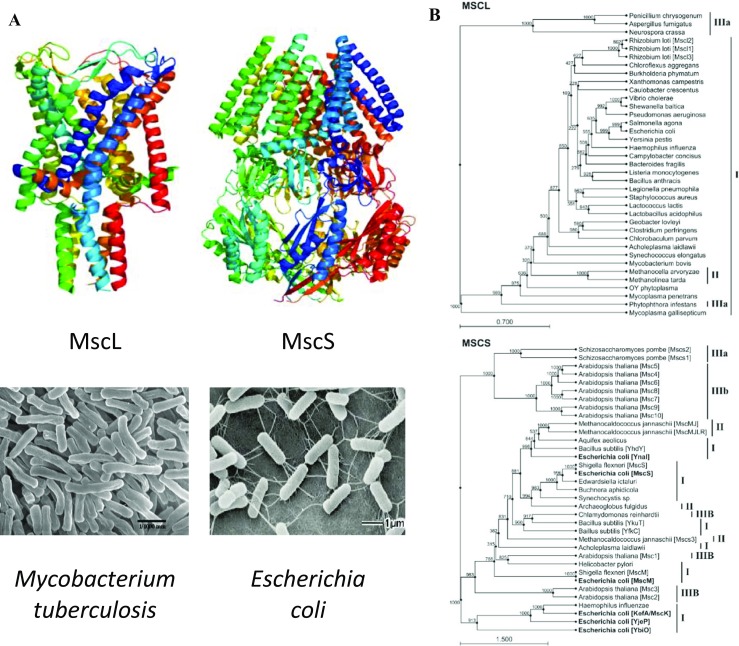
Structure and phylogeny of bacterial mechanosensitive channels. a 3D structure of the MscL pentamer of Mycobacterium tuberculosis (Chang et al. 1998) and MscS heptamer of E. coli (Bass et al. 2002) (top) and scanning electronmicrographs of the corresponding bacterial species (bottom). b A reduced phylogenetic trees showing the distribution of MscL and MscS homologs, which are found across all three domains of life. MscL homologs are found in cell-walled and wall-less bacteria (I), archaea (II), and fungi and oomycetes (belonging to the domain eukaryota, IIIa). MscS homologs have additionally been identified in plants and unicellular green algae (IIIb) with multiple MscS homologs often found in a single organism. The different MscS homologs in E. coli are in bold typeface to highlight the presence of at least three distinct clusters: (i) MscS-like, (ii) MscM-like, and (iii) MscK-like, which suggests that the acquisition of multiple MscS-like homologs might have occurred at a much earlier evolutionary time point than the rare cases in which this has occurred for MscL (e.g., Rhizobium loti). Phylogenetic trees were constructed using 1000 bootstrapped iterations of the UPGMA algorithm in CLC Sequence Viewer (CLC bio, Aarhus Denmark). Scale bar indicates substitutions per site. For simplicity, organisms that contain multiple, uncharacterized MscS-like proteins have had them sequentially designated MscsN, where N begins with the number of characterized homologs plus one and increments. M. jannaschii Mscs3 = Uniprot ID Q58111; Schizosaccharomyces pombe Mscs1 = O74839; S. pombe Mscs2 = O14050 (adapted from (Martinac et al. 2013) with permission)
