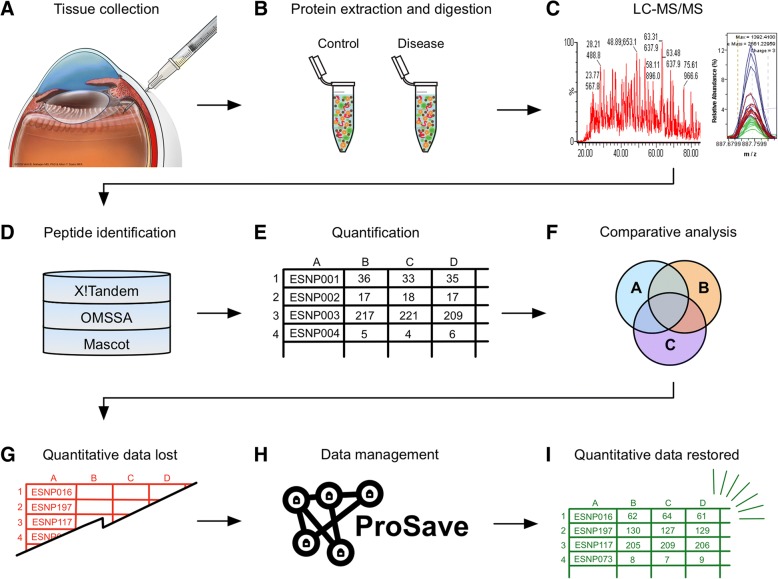
Fig. 1.
Informatics workflow for shotgun proteomics studies: a Liquid biopsies taken at time of surgery. b Liquid biopsies are processed for proteomic analysis. c Liquid chromatography-tandem mass-spectrometry used to analyze protein content. d Protein IDs are matched to peptide mass-spectral data. e Protein IDs and mass-spectra data are organized. f Samples (control vs. disease, etc.) are compared based on protein contents. g Quantitative data is lost during comparative analysis. h ProSave inputs original data and bare protein IDs, then outputs (i) restored protein-data pairs for trend analysis