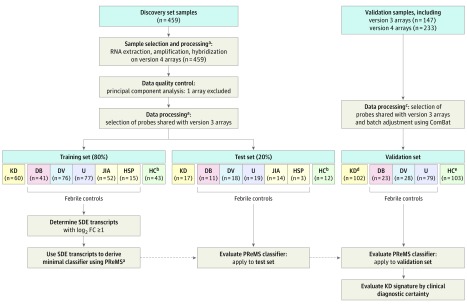
Figure 2. Study Design.
The overall study pipeline shows sample handling, derivation of test and training data sets, data processing, and analysis pipeline. Version 3 arrays indicate HumanHT-12, version 3.0 BeadChip (Illumina); version 4 arrays indicate HumanHT-12, version 4.0 BeadChip (Illumina); and ComBat indicates the ComBat algorithm.23 DB indicates definite bacterial; DV, definite viral; FC, fold change; HC, healthy controls; HSP, Henoch-Schönlein purpura; JIA, juvenile idiopathic arthritis; KD, Kawasaki disease; PReMS, parallel regularized regression model search; SDE, significantly differentially expressed; and U, infections of uncertain bacterial or viral etiology.
aSee Supplemental Methods (RNA sample extraction and processing), as well as Statistical Methods in eMethods in the Supplement.
bHealthy controls were used in model building but were excluded from estimates of model accuracy.
cSee Statistical Methods in eMethods in the Supplement; 146 acute KD samples (HumanHT-12, version 4.0) were used in Combat, of which 101 were taken forward.
dDiagnostic performance was assessed on 72 patients (within the first 7 days of illness).
eIncludes convalescent KD and healthy controls.