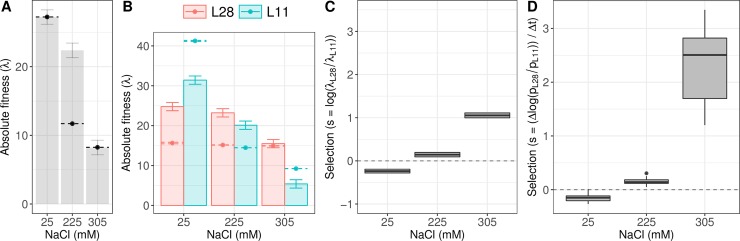
Fig 6. Evaluating the model predictions using fitness data.
(A) Absolute fitness of the ancestor lab-adapted population at three salt concentrations (mean ± SE). The point and dashed error show the expected absolute fitness of the ancestral population, when modelling linear fitness reaction norms of all segregating lineages (from S11 Fig). At 25 mM and 305 mM NaCl the predicted values exactly match the observed values since they were used for inference. (B) Absolute fitness of the lines L28 and L11 at three salt concentrations (mean ± SE), with points and dashed lines as in (A) (from Fig 5A). (C) From (B), estimates (mean ± SE) of the expected relative fitness of L28 to L11 at three salt concentrations. (D) Similar to (C), but estimates from competitive fitness assays between L28 and L11. From ref. [21], the two inferred L28 and L11 lineages were identified after genome-wide sequencing of 100 lines derived from two gradual populations at generation 50 (S13 Fig and S2 Table). For the competitions shown in (D), frequency estimates of L28 and L11 were obtained using pooled-genotyping data on 18 SNPs that differ in L28 and L11 (see S14 Fig for calibration curves on these SNPs, and S15 Fig for our ability to differentiate L28 from L11). Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.76n6f7c [54].