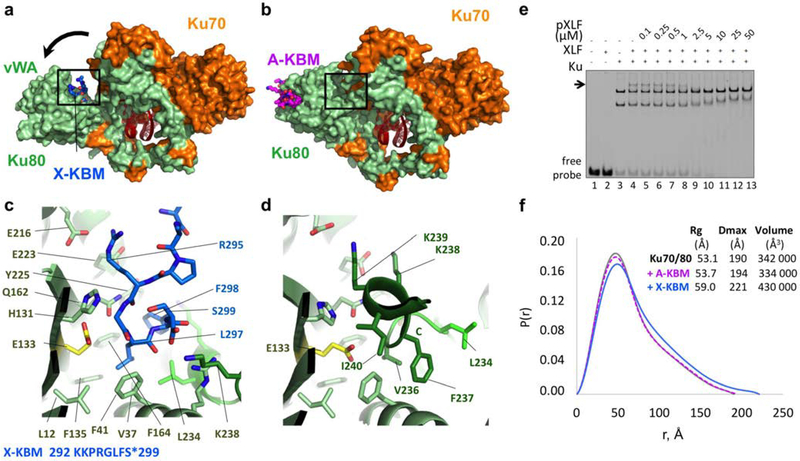
Figure 2. Crystal structure of the XPLF KBM (X-KBM) bound to the Ku80.
(a) Crystal structure of the quaternary complex Ku70-Ku80-DNA-(X-KBM peptide). The X-KBM (blue) binds in an internal site of the Ku80 subunit created upon an outward rotation of the vWA domain. The Ku80 vWA opening creates a large groove between the Ku80 vWA and the rest of the heterodimer. (b) The crystal structure of Ku70/Ku80/DNA in presence of the A-KBM is shown with the same orientation. (c-d) Comparison of the X-KBM binding site in presence of X-KBM (c) or A-KBM (d) peptides. The X-KBM interacts with Ku80 residues involved in Ku intramolecular contacts in the closed state of Ku observed with the A-KBM or with no peptide. The last GLFS residues of the X-KBM interact with the bottom of the groove formed in the open state. The glutamic acid presents an atypical hydrophobic environment and could be at the origin of the vWA instability. The X-KBM residues occupy the position of the helix 236–241 of Ku80 in the closed conformation and some X-KBM side chains (R295X, L297X and F298X) mimic the intramolecular interactions made by Ku80 residues with the vWA domain. (e) Gel shift assay with XLF and Ku in presence of a 50bp DNA with a FAM in 5’and competition with pXLF containing the X-KBM motif. The arrow indicates the XLF Ku-DNA complex. Uncropped gel image is shown in Supplementary Data Set 1. (f) The pair distributions P(r) obtained in solution by SAXS analysis indicates an opening of the Ku70/Ku80/DNA complex with higher Dmax and Rg in presence of the X-KBM (blue line) compared to the Ku/DNA complex without peptide (grey line) and to the A-KBM complex (magenta line). Values deduced from SAXS analysis are reported beside the curves.