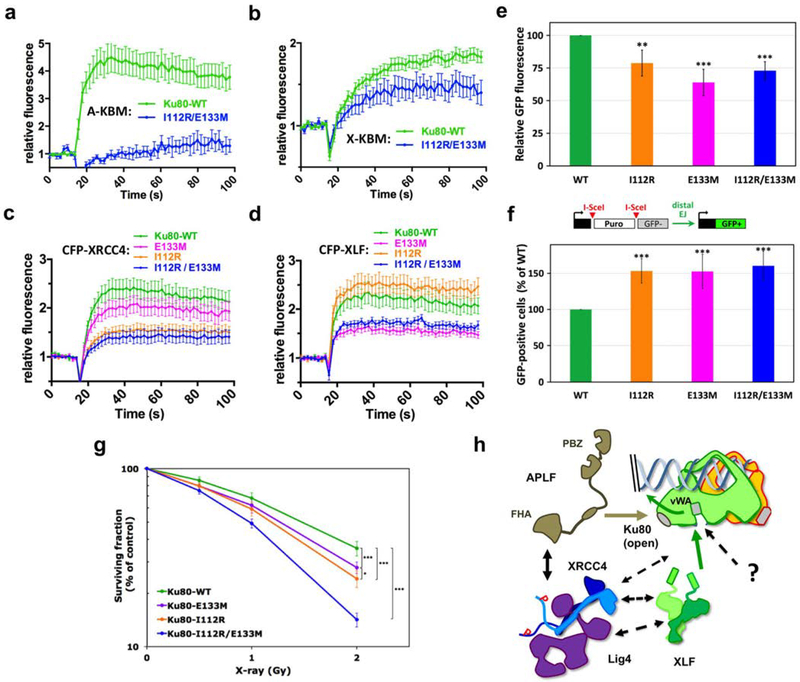
Figure 5. Effects of Ku80 mutations in APLF and XLF binding sites.
(a-b) Dynamics of CFP-(A-KBM) (a) and (X-KBM) (b) at laser damaged sites, in U2OS cells expressing wild-type or I112R/E133M mutant Ku80. n=20 and 9 cells in (a) and n=48 and 11 cells in (b) for WT and mutant Ku80. p values at last time point: (a) WT vs I112R/E133M p=0.001; (b) WT vs I112R/E133M p=0.0111. (c-d) Dynamics of CFP-XRCC4 (c) and XLF (d) in cells expressing wild-type, I112R, E133M or I112R/E133M mutant Ku80. n=38, 27, 28, and 24 cells for WT, E133M, I112R and I112R/E133M Ku80 conditions in (c) and n=24, 26, 20 and 23 cells for I112R, WT, I112R/E133M and E133M Ku80 conditions in (d). p values at last time point: (c) WT vs E133M p=0.532; WT vs I112R p=0.0133; WT vs I112R/E133M p=0.0048; (d) WT vs I112R p=0.246; WT vs E133M p=0.0048; WT vs I112R/E133M p=0.0248. (e) End-joining activity in U2OS cells expressing mutated or WT Ku80. Error bars represent s.d., n=4 independent experiments. p values (unpaired two-tailed t- test): WT vs E133M p=0.0004; WT vs I112R p=0.0052; WT vs I112R/E133M p=0.0002. (f) Distal end-joining in U2OS cells containing mutated or WT Ku80. Error bars represent s.d., n=7 independent experiments. p values (unpaired two-tailed t-test): WT vs E133M p=7.49 e-05; WT vs I112R p=2.21 e-06; WT vs I112R/E133M p=4.05 e-06. (g) Survival of U2OS cells expressing WT or mutated Ku80. y axis is log scale. Error bars represent s.d., n=7 to 10 independent experiments. p (unpaired two-tailed t-test): WT vs I112R p=1.47 e-06; WT vs E133M p=6.32 e-05; WT vs I112R/E133M p=2.52 e-13; I112R vs E133M p=0.011. Significant p-values are indicated as follows: *p< 0.05, **p < 0.01, ***p < 0.001. (h) Model for APLF and XLF KBMs function during NHEJ.