Figure 5. Bioinformatics of shape, applied to planaria.
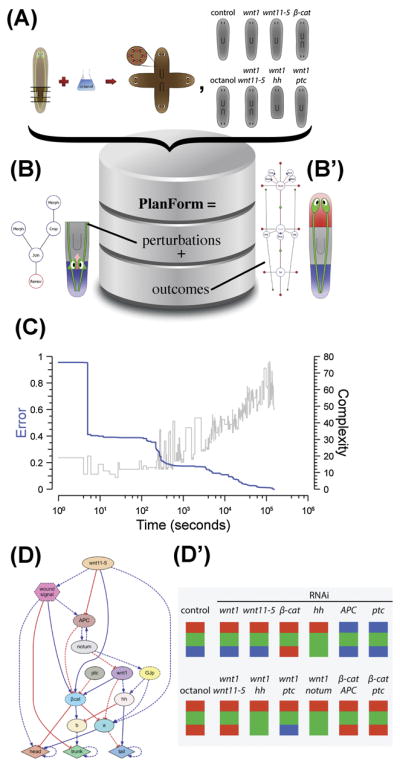
(A, B) The continued development of knowledge in this field will require computational tools going beyond bioinformatics of genes and proteins, to assist in development and analysis of models. One effort, PlanForm [169], comprises over 1,000 experiments from the literature, matching the functional manipulations performed (e.g., specific cuts, joins, RNAi, bioelectric change; see B) and the resulting anatomical outcomes represented by a graph notation (B′).
(C) One recent application of artificial intelligence to discovery of regulator pathways [168] used evolutionary selection over a population of biochemical models. Here shown as the progressive reduction of error in the predictions of top candidate models at each generation.
(D) This process uncovered a gene regulatory network whose patterning properties matched observed data on canonical pathways (a sample is shown in D′).
