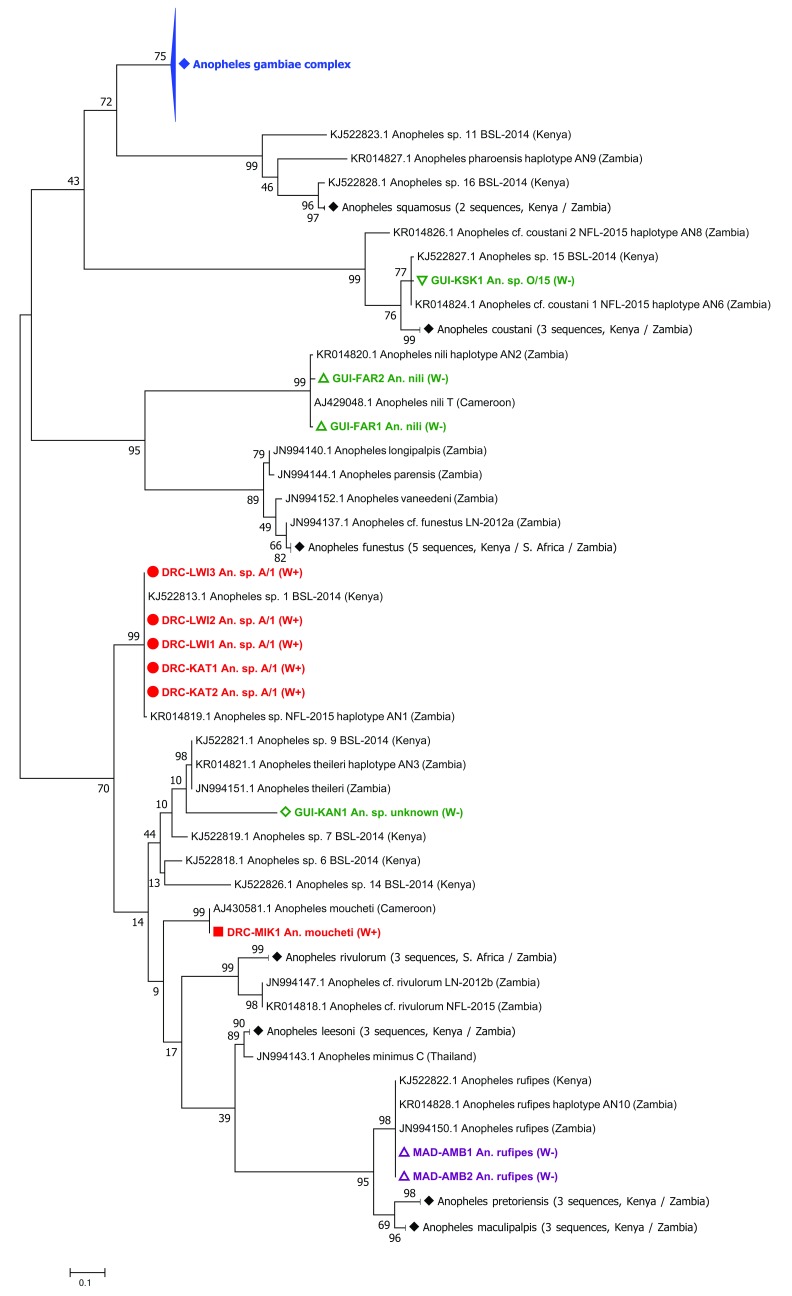
Figure 3. Maximum Likelihood molecular phylogenetic analysis of Anopheles ITS2 sequences from field-collected mosquitoes outside of the An. gambiae s.l. complex.
The tree with the highest log likelihood (-3084.12) is shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 118 nucleotide sequences. There were a total of 156 positions in the final dataset. Symbols, colours and codes used for sequences generated in this study are as follows: W+; individual was Wolbachia positive (solid coloured symbol), W-; individual was Wolbachia negative (empty coloured symbol). DRC, Democratic Republic of the Congo (red): KAT, Katana; LWI, Lwiro; MIK, Mikalayi. GUI, Guinea (green); FAR, Faranah; KAN, Kankan; KSK, Kissidougou. MAD, Madagascar (purple); AMB, Ambomiharina. Different shape coloured symbols are used to differentiate between different mosquito species. GenBank sequences included (for comparison with sequences generated in this study) are in black with their accession numbers provided. Where GenBank sequence subtrees have been compressed, this is denoted by a solid black diamond symbol. GenBank accession numbers for sequences included in compressed subtrees are as follows: Anopheles squamosus; KJ522825.1 and KR014825.1. Anopheles coustani; JN994134.1, KJ522815.1 and KR014823.1. Anopheles funestus; AF062512.1, JN994135.1, JN994136.1, KJ522816.1 and KR014830.1. Anopheles rivulorum; JN994148.1, JN994149.1 and KR014822.1. Anopheles lessoni; JN994139.1, KJ522824.1 and KR014834.1. Anopheles pretoriensis; JN994145.1, KJ522820.1 and KR014829.1. Anopheles maculipalpis; JN994142.1, KJ522817.1 and KR014835.1. (The blue Anopheles gambiae complex compressed subtree is shown in Figure 2.)