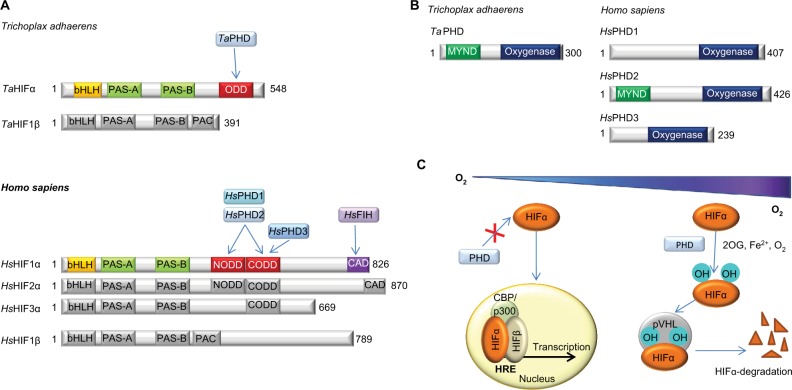
Figure 1.
Overview of the HIF system.
Notes: (A) Comparison of domain architectures of HIFα and HIFβ in T. adhaerens and humans. In contrast to the multiple PHD/HIFα-isoforms and ODDs present in vertebrates, the prime components of the HIF system in T. adhaerens involve only one PHD and one HIFα, which has a single ODD.16 Arrows indicate assigned prolyl and asparaginyl hydroxylation sites. (B) Domain structures of HIF prolyl hydroxylases (PHDs) in T. adhaerens and humans. Domain acronyms: 2OG dioxygenase domain (oxygenase), MYeloid, Nervy, and DEAF-1 (MYND)-type zinc finger domain (MYND). (C) Outline of the conserved mechanism of the response to chronic hypoxia in animals. The HIF transcription factors are regulated by PHD catalyzed hydroxylation of prolyl residues in the ODD(s) of HIFα under normoxia. Recognition of the hydroxylated prolyl residues by pVHL is followed by ubiquitination by the E3 ubiquitin ligase, which tags HIFα for proteasomal degradation. In humans, FIH, which is only sporadically present in non-vertebrate animals,16 constitutes an additional oxygen dependent regulatory element of HIF activity. FIH catalyzes the hydroxylation of an asparagine residue in the CTAD of HIFα, thus disrupting its interaction with the CBP/p300 transcriptional co-activator, and hindering transcriptional activation.11
Abbreviations: PHD, Prolyl hydroxylases; HIF, hypoxia-inducible transcription factor; ODD, oxygen-dependent degradation domain; bHLH, basic helix-loop-helix motif; PAS, Per-ARNT-Sim domain; CTAD, C-terminal transactivation domain; PAC, PAS-associated C-terminal domain; FIH, factor inhibiting HIF.