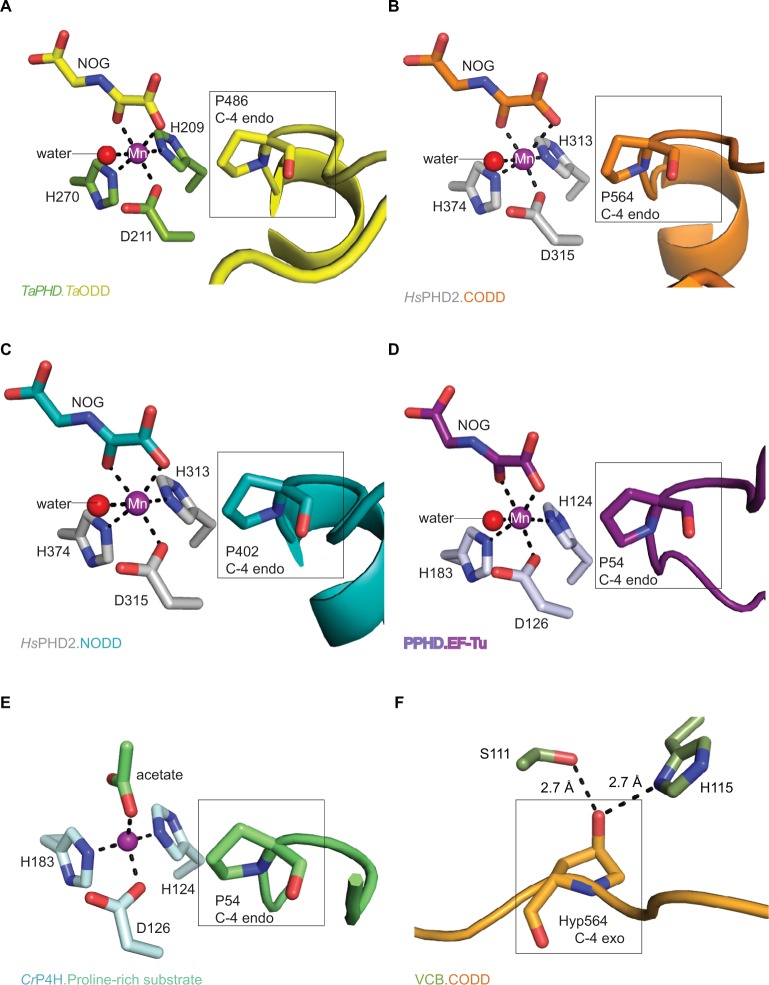
Figure 6.
Active site metal region details and comparison of the conformations of the target prolyl-residues in the enzyme-substrate complex structures of TaPHD, HsPHD2, PPHD, and CrP4H.
Notes: (A) TaPHD.TaODD, (B) HsPHD2.CODD (PDB: 3HQR,34), (C) HsPHD2.NODD (PDB: 5L9V,34), (D) PPHD.EF-Tu, (PDB: 4IW3,38) and (E) CrP4H.proline-rich substrate (PDB: 3GZE,66) complex structures. Note that, in all the enzyme-substrate complexes, the proline C-4- methylene, that is hydroxylated, adopts the endo-conformation. (F) By contrast, Hyp564 in HsHIF1α CODD adopts the C-4 exo-conformation when bound to the VCB complex (PDB: 1LM8,55–57). In (A and D), N-oxalylglycine (NOG) acts as a 2OG analog. Note that the Zn (substituting for Fe) in the CrP4H active site is tetrahedrally coordinated, with an acetate binding instead of the 2OG co-substrate. The structures reveal conserved orientations of the “target” proline residues, which in each case, adopt a C-4 endo-conformation.55 Note that the metal bound water present in the TaPHD (and HsPHD2/PPHD) substrate complex structures is not observed in the CrP4H.(Ser-Pro)5 structure.