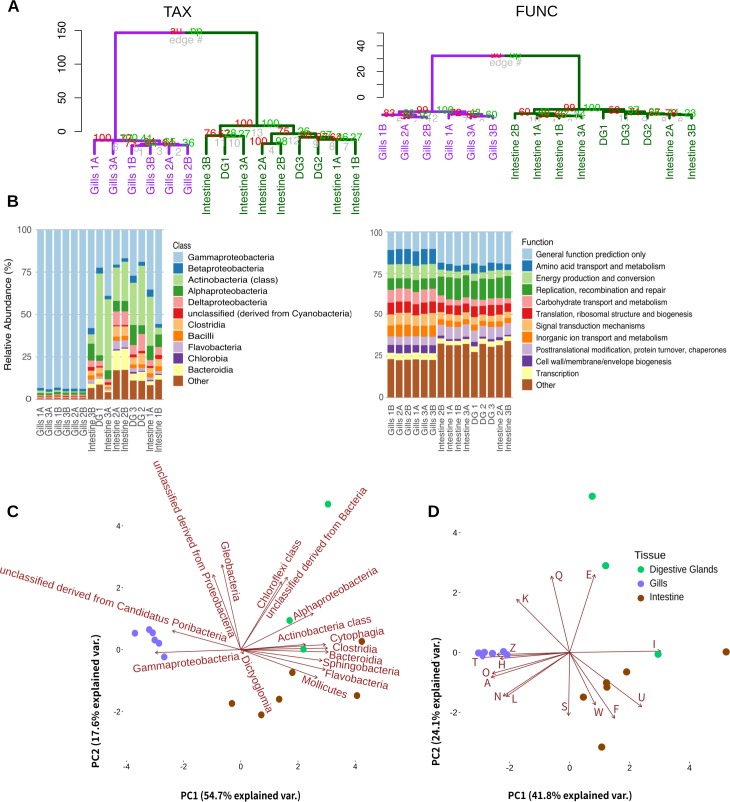
Fig 1. Multivariate analysis of metagenomes microbial taxonomical and functional features.
A) Unsupervised hierarchical clustering of taxonomical (TAX) and functional (FUNC) features. Clades formed by gill samples and intestine plus digestive gland samples are highlighted respectively in purple and dark green. The pvclust AU (Approximately Unbiased) and BP (Bootstrap Probability) p-values are shown at clades nodes in red and light green respectively. B) Relative abundances of metagenome bacterial classes and functions respectively signed with RefSeq and COG (Level 2) databases. C) Multiple dimensional scale (MDS) plot of N. reynei metagenomics samples using distances and the fifteen most important bacteria classes (RefSeq database) and functions (COG database at Level 2) vectors as calculated from unsupervised random forest. D) Digestive gland, intestine and gill samples are shown as green, brown and purple dots respectively. Clusters of Orthologous Groups are classified into functional categories as follow: A—RNA processing and modification; E—Amino acid transport and metabolism; F—Nucleotide transport and metabolism; H—Coenzyme transport and metabolism; I—Lipid transport and metabolism; K–Transcription; L—Replication, recombination and repair; N—Cell motility; O—Post-translational modification, protein turnover, and chaperones; Q—Secondary metabolites biosynthesis, transport, and catabolism; S—Function Unknown; T—Signal transduction mechanisms; U—Intracellular trafficking, secretion, and vesicular transport; W—Extracellular structures; Z–Cytoskeleton.