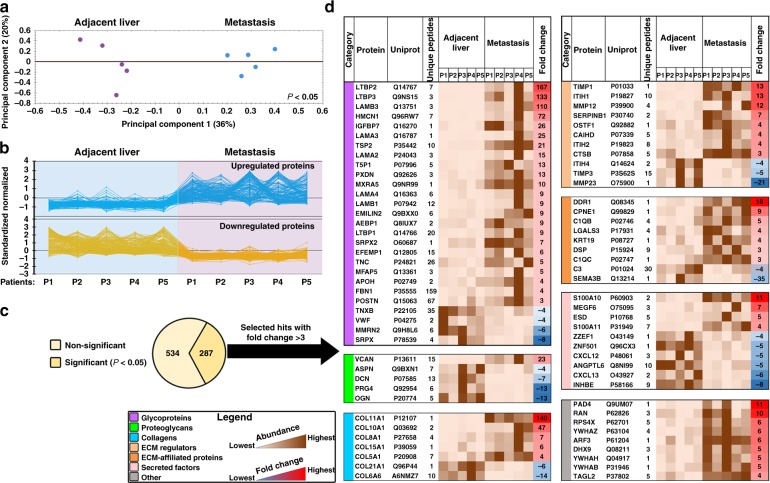
Fig. 1.
Label-free quantitative proteomics identifies the distinctive ECM composition of human CRC hepatic metastases. a Decellularized and ECM-enriched fractions from CRC hepatic metastases and paired adjacent unaffected liver tissues (n = 5 per group) were analyzed using LC-MS/MS with the following label-free quantitation. Shown is the principal component analysis of the relative protein abundance between metastasis and adjacent liver groups (P < 0.05). Percentage of variance is shown in parentheses. b Clustering analysis graph reveals categories of proteins differentially expressed in metastasis and adjacent liver groups. c Statistically significant differentially expressed proteins (287 out of 821 hits, two-way ANOVA, P < 0.05) were further classified either as ECM proteins or other proteins. d A heat map illustrating significantly different matrisome proteins (69 hits), which were selected based on a fold change threshold of > 3 and ranked in accordance with ECM category and fold change. The last gray colored table represents 9 selected other hits (i.e., non-ECM proteins yet significantly different in ECM-enriched samples). ECM extracellular matrix, P patient