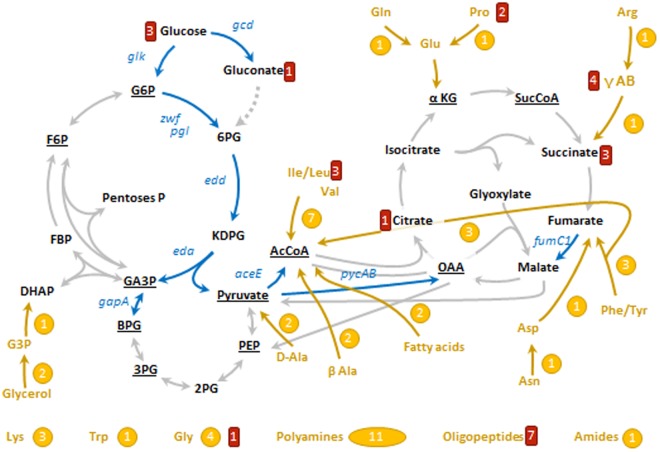
Figure 4.
Targets of Crc on bacterial metabolism and on the transport of carbon sources. The diagram represents schematically the main pathways of central carbon metabolism, some pathways of carbon sources catabolism, some of which elements are regulated by Crc, and transporters repressed by Crc. In black, key metabolites within central carbon metabolism, underlined the 12 precursors of biomolecules30. In blue, genes encoding enzymes of central carbon metabolism which translation repress the protein Crc (PTV > 1), including Zwf (glucose-6-P 1-dehydrogenase), PgI (6-phosphogluconolactonase), Edd (6-phosphogluconate dehydratase), Eda (KDPG aldolase), Glk (glucokinase), Gcd (glucose dehydrogenase), GapA (GA3P dehydrogenase), AceE (pyruvate dehydrogenase), PycAB (pyruvate carboxylase), and FumC1 (fumarase). In grey, other pathways of central carbon metabolism that are not controlled by Crc. In yellow, secondary carbon sources which degradation is controlled by Crc, including a simplified pathway indicating its connection to central carbon metabolism and the number of enzymes which production is controlled by Crc. In red, the number of proteins related with the transport of such carbon source. This scheme has been done with the information of the databases Pseudomonas Genome database61, MetaCyc62 and TransportDB63 and the information of the bibliography, specially refs38,64. G6P, glucose-6-P; F6P, fructose-6-P; FBP, fructose-1,6-P2; GA3P, glyceraldehyde-3-P; DHAP, dihydroxyacetone-P; BPG, 1,3-biphosphoglycerate; 2PG, 2-P-glycerate; 3PG, 3-P-glycerate; PEP, phosphoenolpyruvate; AcCoA, acetyl-coenzyme A; 6PG, 6-P-gluconate; KDPG, 2-keto-3-deoxy-6-P-gluconate; αKG, α-ketoglutarate; SucCoA, succinyl-coenzyme A; OAA, oxaloacetate; E4P, erythrose-4-P; R5P, ribulose-5-P; G3P, glycerol-3-P; D-Ala, D-alanine; β-Ala, β-alanine; γ-AB, γ-aminobutyrate.