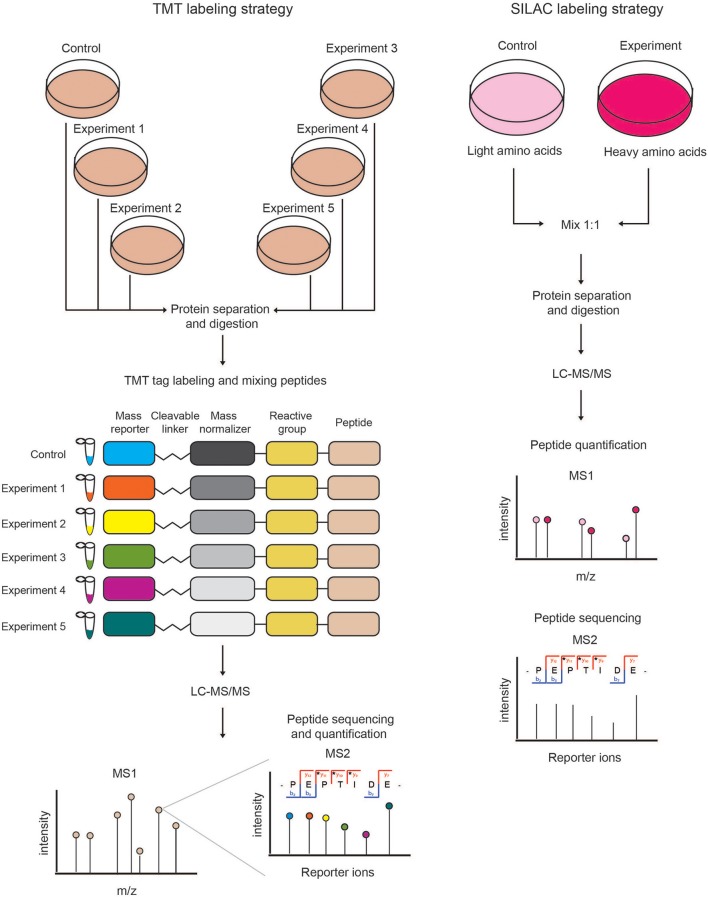
Figure 2.
Schematic view of chemical (TMT) and metabolic (SILAC) quantitative proteomics strategies. (Left) In the TMT labeling strategy, protein extracts derived from different samples are reduced, alkylated, Trypsin-digested and then in vitro labeled using the isobaric TMT tags. The resulting peptides are mixed in equal amounts and analyzed by LC-MS/MS. At the MS1 level, the isobaric peptides appear as a single precursor ion, while at the MS2 level the different reporter ions are separated according to their mass. In this approach, peptides are identified and quantified at the MS2 level. (Right) In SILAC labeling strategy, cells are metabolically labeled by growing them in medium containing either light or heavy amino acids. Cells from the two experimental conditions are harvested, mixed in equal amounts and lysed to obtain a protein extract that is then reduced, alkylated and Trypsin-digested. The resulting peptides are analyzed by LC-MS/MS. Each protein-derived peptide appears as a peak-pair at the MS1 level, whereby the heavy and light peptides will be distinguishable according to their nominal mass. In metabolic labeling approaches, peptides are quantified and identified at the MS1 and MS2 levels, respectively.