Figure 1. Structure and biochemical characterization of the C‐terminus of SPIN90.
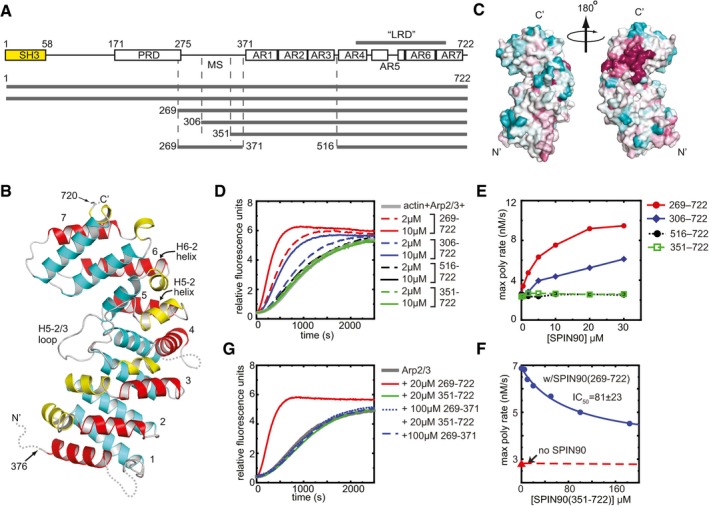
- Domain organization of SPIN90 and SPIN90 constructs used in this study (SH3, Src homology 3; PRD, proline‐rich domain; MS, middle segment; AR, armadillo repeat).
- Ribbon diagram of SPIN90 (351–722). Helices are colored according to their position within the repeat (1—yellow, 2—red, 3—cyan). Armadillo repeats are numbered 1–7. Disordered residues are indicated with dotted lines.
- Surface representation of SPIN90 (351–722) showing conservation scores derived from an alignment of WDS family proteins from 31 diverse species (see Materials and Methods). Conservation score is indicated by color, with dark red > pink > light pink > gray > light cyan > cyan, and higher scores meaning greater conservation.
- Time course of pyrene actin polymerization showing the influence of different SPIN90 constructs in reactions containing 50 nM Bos taurus Arp2/3 complex and 3 μM 15% pyrene‐labeled actin.
- Plot of maximum polymerization rate versus concentration of SPIN90 for reactions shown in (D).
- Plot of maximum polymerization rate versus concentration of SPIN90 for pyrene actin polymerization assays containing 2.5 μM SPIN90, 500 nM Arp2/3 complex, 3 μM 15% pyrene‐labeled actin and the indicated concentration of the SPIN90 ARM domain construct.
- Time courses of pyrene actin polymerization as described in panel (D), except the influence of the SPIN90 middle segment is tested on its own or in the presence of the 351–722 construct.
