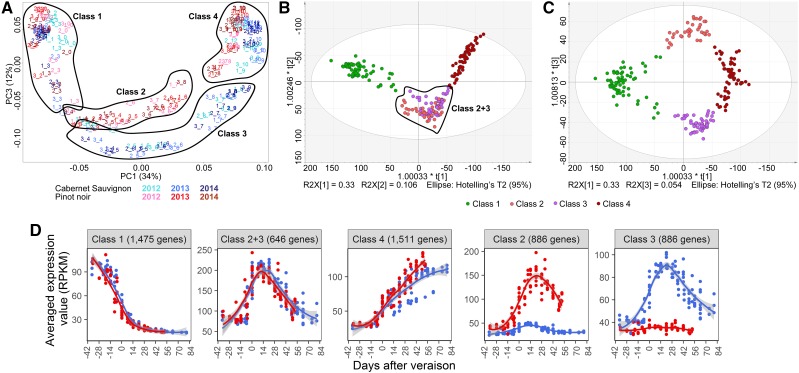
Figure 4.
PCA showing the distribution of gene expression in Cabernet Sauvignon and Pinot Noir berries for all three vintages. A, PC1 and PC3 explain 46.2% of the variability. PC1 represents the distribution of samples according to time and/or development for both varieties. The early and late developmental time points cluster together (circled and labeled Class 1 and Class 4, respectively). PC3 reveals the transcriptional programs of berry maturation, highlighting both universal genetic control (Classes 1 and 4) as well as varietal-specific programs (circled and labeled Class 2 and Class 3). B, O2PLS-DA distribution of the same samples to verify the four-class clustering. R2X[1] and R2X[2] show the clusters describing immature/preveraison berries (Class 1), mature berries (Class 4), and non-varietal-specific development (Classes 2+3). C, R2X[1] and R2X[3] show the clusters describing immature/preveraison berries (Class 1), mature berries (Class 4), and varietal-specific development (Pinot Noir, Class 2; Cabernet Sauvignon, Class 3). D, Averaged expression profile of the five gene classes show consistent and distinguishable trends across fruit development and variety. Line graphs were created using data from three vintages plotted by days after veraison. Gray shading indicates 0.95 confidence levels relative to the smoothed conditional means plotting method.