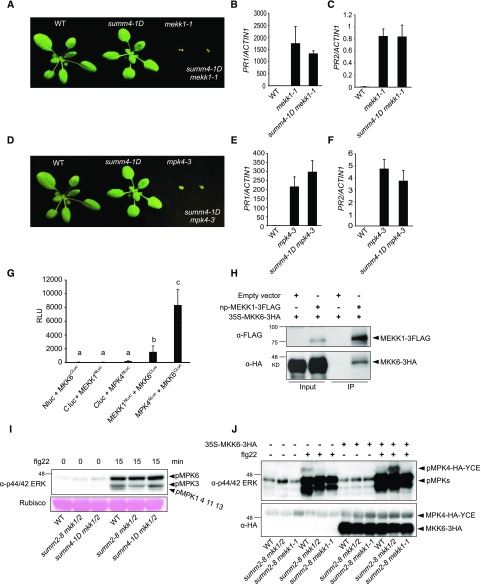
Figure 3.
MEKK1, MKK6, and MPK4 function in a MAPK cascade. A, Morphology of 3-week-old wild-type (WT), summ4-1D, mekk1-1, and summ4-1D mekk1-1 plants. B and C, Expression levels of PR1 (B) and PR2 (C) in wild type, mekk1-1, and summ4-1D mekk1-1. Values were normalized relative to the expression of ACTIN1. Error bars represent standard deviations from three measurements. D, Morphology of 3-week-old wild-type, summ4-1D, mpk4-3, and summ4-1D mpk4-3 plants. E and F, Expression levels of PR1 (E) and PR2 (F) in wild-type, mekk1-1, and summ4-1D mekk1-1. Values were normalized relative to the expression of ACTIN1. Error bars represent standard deviations from three measurements. G, Interactions between MKK6 and MEKK1 or MPK4. Luciferase activities from split luciferase complementation assays represented in relative light units (RLUs). Error bars represent standard deviations from eight replicates. Statistical differences among the samples are labeled with different letters (P < 0.05, one-way ANOVA/Tukey’s test, n = 8). H, Analysis of the interaction between MKK6-3HA and MEKK1-3FLAG by coimmunoprecipitation. The proteins were transiently expressed in N. benthamiana using Agrobacteria strains carrying 35S-MKK6-3HA or a native promoter driven MEKK1-3FLAG (np-MEKK1-3FLAG). Immunoprecipitation was carried out using anti-FLAG beads. Western blot was carried out using anti-FLAG or anti-HA antibodies. I, MAPK activation in wild type (WT), summ2-8 mkk1/2, and summ4-1D mkk1/2. Plants were sprayed with 100 nm flg22. Samples were harvested at 0 and 15 min after flg22 treatment. MAPK activation was detected by immunoblotting with antip44/42 ERK antibody. Input was visualized by Ponceau S staining of Rubisco. J, The 35S-MPK4-HA-YCE construct was cotransfected with 35S-MKK6-3HA into protoplasts isolated from wild-type, summ2-8 mkk1/2, and summ2-8 mekk1-1. The samples were treated with or without 100 nm flg22 for 15 min before they were collected for western blot analysis using antip44/42 ERK or anti-HA antibodies.