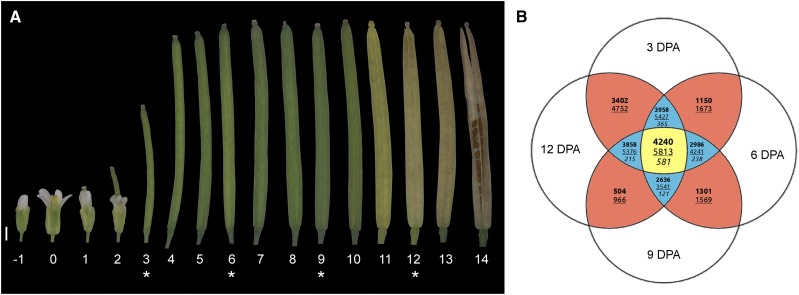
Figure 1.
RNA sequencing (RNAseq) strategy used to explore the mechanisms controlling fruit formation and maturation. A, Developmental series of Arabidopsis siliques from the flower bud (−1 DPA) to the mature fruit (14 DPA). Numbers below the siliques indicate the DPA. The siliques at 3, 6, 9, and 12 DPA, marked with asterisks, were opened and the seeds were removed prior to use for RNAseq analysis. The image shown is a composite image. Bar = 1 mm. B, Venn diagram shows the number of up-regulated (boldface), down-regulated (underlined), and alternative behavior (italics) genes across all comparisons performed using the selected cutoff (fold change of 2.5 and P = 0.001; see “Materials and Methods”). The four pairwise comparisons (6 versus 3 DPA, 9 versus 6 DPA, 12 versus 9 DPA, and 12 versus 3 DPA) show only two values, since, by definition, genes can only be either up-regulated or down-regulated. The center of the Venn diagram (i.e. the yellow portion) shows the number of differentially expressed genes considered in this work: 4,240 up-regulated genes (Supplemental Table S1), 5,813 down-regulated genes (Supplemental Table S2), and 581 genes that showed an alternative behavior (Supplemental Table S3–S6).