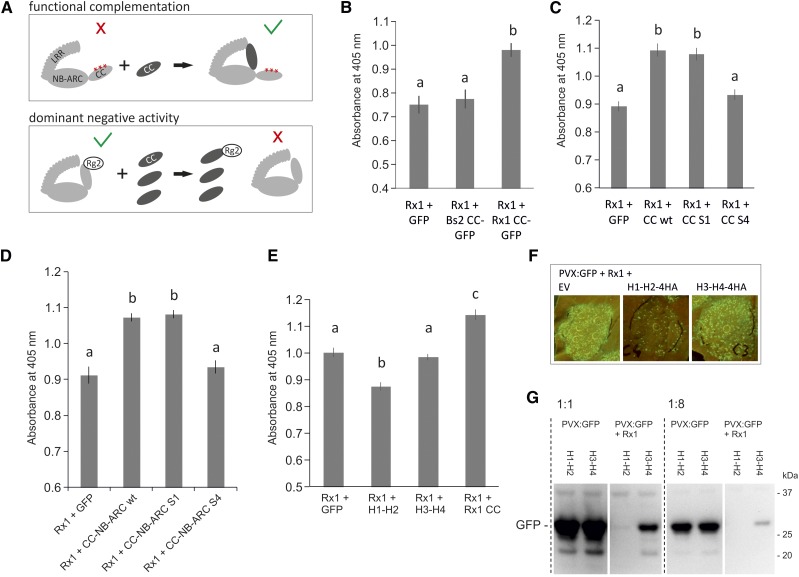
Figure 8.
Exploring suppressive effects of the coexpressed CC domain on the functionality of wild-type Rx1. A, Schematic view of the potential mechanisms behind the functional complementation and the dominant negative phenotypes that could occur upon coexpression of the wild-type CC domain with mutant or wild-type Rx, respectively. A red cross indicates a loss of signaling of the full-length protein, and the green V indicates a state in which the protein is able to signal. B, Assay to test if coexpression of the CC of Rx1 suppresses the resistance mediated by Rx1, leading to higher PVX levels in a transient PVX resistance assay. Coexpression of GFP or the CC domain of Bs2 was used as negative controls. PVX accumulation was determined via an anti-PVX ELISA. Error bars present the se (n = 8), and letters denote significantly (P < 0.05) different groups (one-way ANOVA with posthoc Tukey’s test). C, To determine if the S1 or S4 mutation affected the suppressive effect seen for the wild-type Rx1 CC, CC constructs harboring these mutations were coexpressed with wild-type Rx1 in a transient PVX assay. Coexpression of GFP served as a negative control. Error bars present the se (n = 12), and letters denote significantly (P < 0.05) different groups (one-way ANOVA with posthoc Tukey’s test). D, Coexpression of the wild type and S1 and S4 variants of the CC-NB-ARC with wild-type Rx1 in a transient PVX assay to test if the mutations affect the suppressive effect of the CC-NB-ARC. Coexpression of GFP was used as a negative control. Error bars present the se (n = 8), and letters denote significantly (P < 0.05) different groups (one-way ANOVA with posthoc Tukey’s test). E, Coexpression of the N- and C-terminal CC segments with the full-length Rx1 in a transient PVX resistance assay to test if the suppressive effect is caused by a specific region in the CC. Error bars present the se (n = 8), and letters denote significantly (P < 0.01) different groups (one-way ANOVA with posthoc Tukey’s test). F, Coexpression of PVX:GFP, Rx1, and the CC segments. The virus accumulation was visualized via the GFP expressed from the PVX genome. Lower GFP levels are apparent on leaves in which H1-H2 is coexpressed. G, To test if the reduction of PVX levels was a direct effect of the H1-H2 segment on the virus or if it was due to an enhancement of Rx1 activity, the CC segments were coexpressed with PVX:GFP in the absence and presence of Rx1. PVX levels were compared on an anti-GFP immunoblot via the GFP expressed by PVX. All samples were loaded undiluted and 8× diluted to make a better comparison possible between the samples with high and low GFP concentrations.