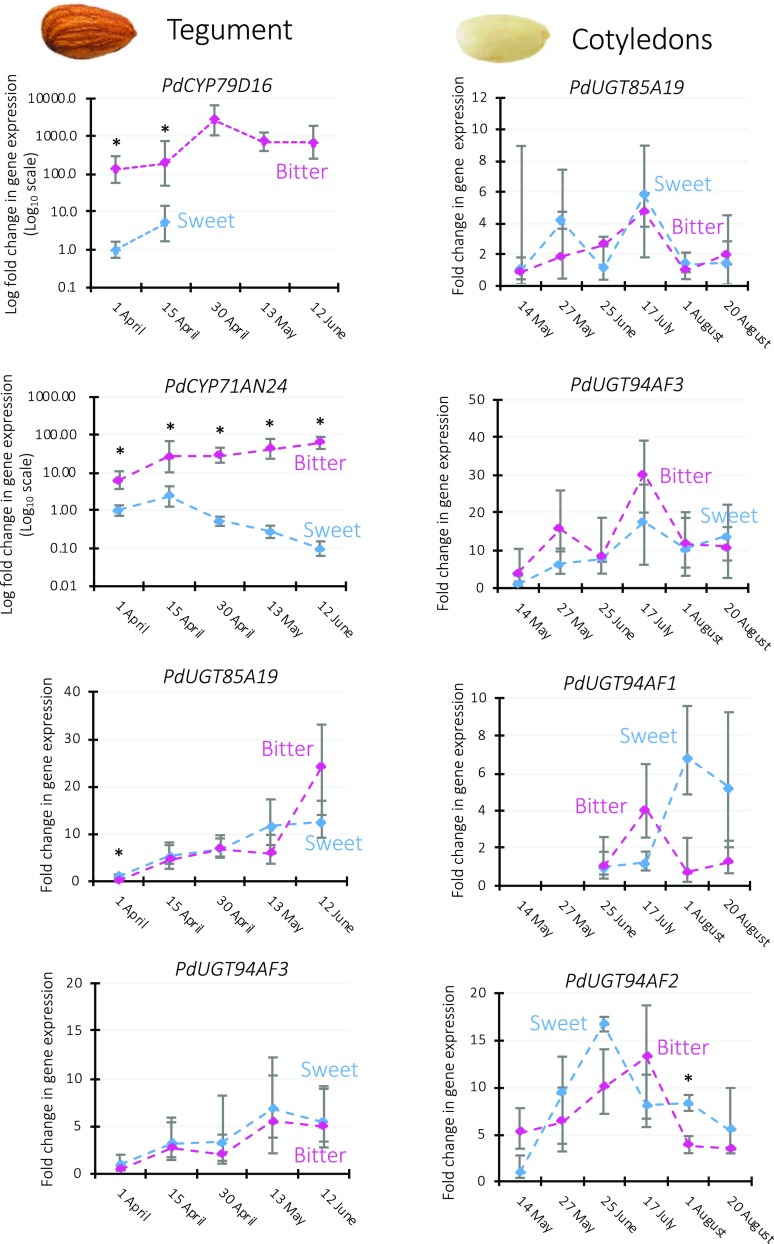
Figure 3.
Relative transcript levels of prunasin and amygdalin biosynthetic genes in the tegument and cotyledons of the sweet genotype Lauranne (blue traces) and the bitter genotype S3067 (purple traces) during fruit development. For each gene, expression levels were normalized to the one displayed by the sweet genotype at the earliest time point. TEFII, RPTII, and UBQ10 were used as housekeeping reference genes. Data are presented as means ± sd of three biological replicates. Note the logarithmic scale applied in the two graphs showing PdCYP79D16 and PdCYP71AN24 in tegument. Asterisks indicate statistically significant genotypic differences at the same time point (Student’s t test, P < 0.01). The expression of PdCYP79D16 was not detected in the sweet tegument at the last three time points, while the expression of PdUGT94AF1 was not detected in the bitter cotyledons at the first two time points. No expression of PdUGT94AF1 and PdUGT94AF2 in tegument and of PdCYP79D16 and PdCYP71AN24 in cotyledons was detectable. For relative expression level comparisons among the enzymes in the same tissue, see Supplemental Figure S5.