Figure 6: Forward simulations enable inferences about negative selection and rare variant architectures.
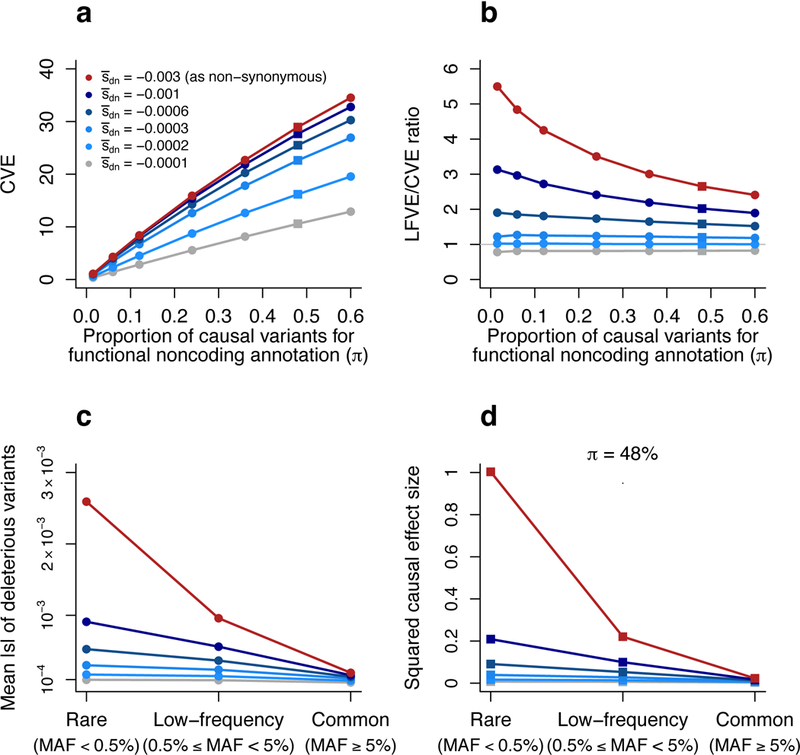
Results are based on forward simulations involving an annotation mimicking functional noncoding variants, as well as other annotations (see text). (a,b) We report the CVE (a) and LFVE/CVE ratio (b) of the functional noncoding annotation as a function of the mean selection coefficient for de novo deleterious variants () and the probability of a de novo variant to be causal (π) for this annotation. and π values for non-synonymous and ordinary noncoding annotations are described in the main text. (c) We report the mean absolute selection coefficient of deleterious variants in the functional noncoding annotation as a function of and MAF (rare, low-frequency, common). (d) We report the mean squared per-allele effect size of causal variants in the functional noncoding annotation (normalized by the mean squared per-allele effect size of rare causal non-synonymous variants) as a function of and MAF (rare, low-frequency and common). Red lines denote the value =−0.003 used to simulate non-synonymous variants, grey lines denote the value =−0.0001 used to simulate ordinary noncoding variants (see main text). The value π=48% used in (d) (see Methods) is denoted via squares in (a) and (b). Numerical results are reported in Supplementary Table 12.
