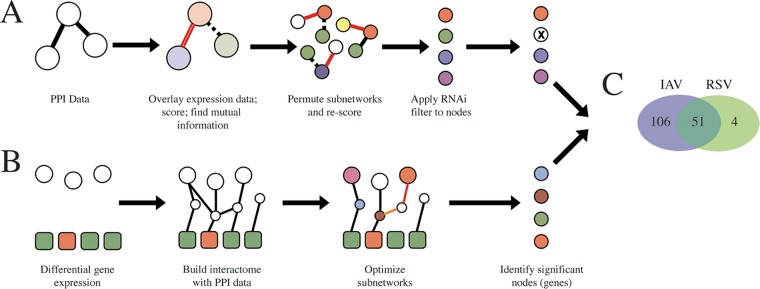
FIG 2.
Schematic overview of the network analyses performed to generate the targeted list of host factors required for viral replication. (A) In the Mutual Information method, an interactome was assembled using protein-protein interaction data (empty circles), and gene expression data were overlaid (colored circles). Each subnetwork was scored by averaging the normalized expression values, and the discriminative potential of each subnetwork was determined based on the mutual information corresponding to its score and the class label of the expression data (i.e., infected versus mock). To identify significant subnetworks, their discriminative power was compared to that of randomly permuted subnetworks, retaining higher-scoring subnetworks. Finally, genes in significant subnetworks were filtered using the aggregate list of previously published RNAi hits from IAV (this step was not performed for RSV). (B) The SteinerNet method attempts to link RNAi screen hits (empty circles) from König et al. (10) to differential expression data (either mRNAseq or microarray data; colored squares) using a protein-protein interaction network. First, an interactome is assembled using PPI data. The subnetworks are mathematically optimized, and the genes in the optimal subnetworks are enriched for their requirement in viral replication. Due to the requirement for RNAi data, this analysis was only performed for influenza A virus. (C) The data from the two network analyses were aggregated, and results are shown in a Venn diagram. A total of 51 genes had shared support for IAV and RSV, while 106 and 4 had support for only IAV and only RSV, respectively.