Figure 1. Structural insights into a 5-HT2B activation mechanism.
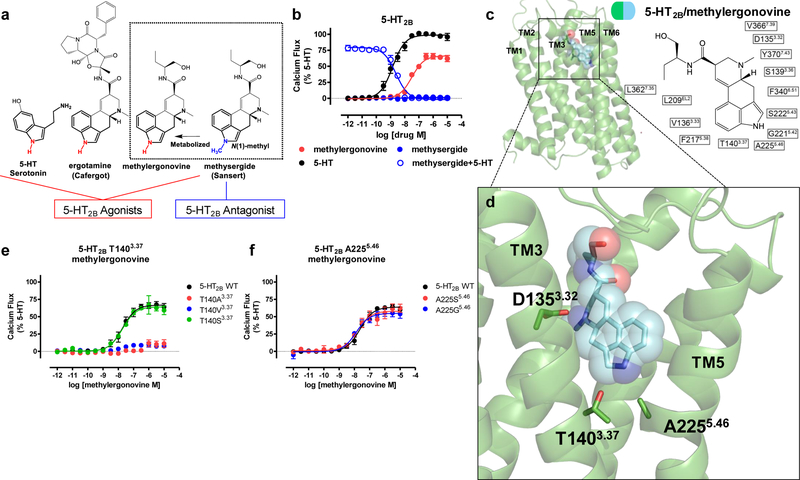
Identification of an 5-HT2BR activation mechanism by the 5-HT2BR-methylergonovine structure a) Structure-activity-relationship of 5-HT2BR ergoline ligands comparing unsubstituted N(1)-H ligands (red) such as 5-HT, ergotamine, and methylergonovine to N(1)-methyl methysergide (blue) b) 5-HT2BR Gq calcium flux activity by 5-HT (black, EC50 = 1.4 nM, Emax = 100%), methylergonovine (red, EC50 = 31 nM, Emax = 66%) and lack of agonist activity by methysergide (blue, closed circles). Methysergide instead acts as a competitive antagonist (blue, open circles, IC50 = 2.4 nM) in response to 5-HT. c) Structure of methylergonovine (blue) at 5-HT2BR (green) with 2D ligand plots of nearby residues (PDB code: 6DRY). d) Close up of methylergonovine binding pose in the binding pocket highlighting D1353.32 interacting with the charged nitrogen of methysergide, and the indole N(1)-H interacting with both T1403.37 and A2255.46 in the orthosteric binding pocket. e) Methylergonovine Gq-mediated calcium flux agonist activity at T140A3.37 (red), T140V3.37 (blue), T140S3.37 (green, EC50 = 18 nM, Emax = 64%) relative to 5-HT2BR wild-type (WT, black, EC50 = 19 nM, Emax = 66%). f) Methylergonovine Gq-mediated calcium flux agonist activity at A225S5.46 (red, EC50 = 15 nM, Emax = 58%), A225G5.46 (blue, EC50 = 12 nM, Emax = 55%) relative to 5-HT2BR wild-type (WT, black, EC50 = 23 nM, Emax = 64%). Data in panels b, e, and f represent mean and S.E.M from three independent experiments (N=3) performed in triplicate.
