Extended Data Table 1:
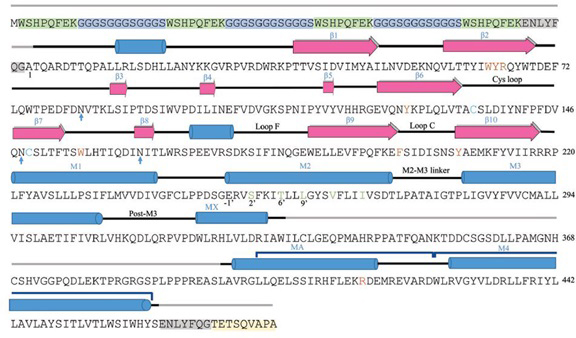
Sequence of mouse 5-HT3AR used in the cryo-EM study and the data on cryo-EM and refinement. a, Full length mouse 5-HT3AR sequence used in the cryo-EM study. Regions in the sequence highlighted in green, blue, gray, and yellow represent strep-tag, linker, TEV cleavage site, and 1D4-tag, respectively. Secondary structural elements as seen in State 1 are plotted above the sequence. Loops in gray color are not seen in the final refined structure. All the important loops, sheets, and helices are labeled. Glycosylation sites are marked as blue arrows. Key residues within the serotonin-binding sites are highlighted in brown color. Cysteines present in the cys-loop are shown as cyan color. Pore-facing residues in M2 are shown in green color. Arg416 in the ICD is shown in red. b, Cryo-EM data collection, refinement and validation statistics.
| a |
 |
||
|---|---|---|---|
| b | State 1 (EMDB-7882) (PDB 6DG7) | State 2 (EMDB-7883) (PDB 6DG8) | |
| Data collection and processing | |||
| Magnification | 130,000x | ||
| Voltage (kV) | 300 | ||
| Electron exposure (e-/Å2) | 40 | ||
| Defocus range (µm) | −1.0 to −2.5 | ||
| Pixel size (Å) | 0.532 | ||
| Symmetry imposed | C5 | ||
| Initial particle images (no.) | 749,970 | ||
| Final particle images (no.) | 103,698 | 18,839 | |
| Map resolution (Å) | 3.32 | 3.89 | |
| FSC threshold | 0.143 | 0.143 | |
| Refinement | |||
| Initial model used (PDB code) | 6BE1 | 6BE1 | |
| Model resolution (Å) | 4.31 | 4.31 | |
| FSC threshold | 0.143 | 0.143 | |
| Map sharpening B factor (Å2) | −50 | −50 | |
| Model composition | |||
| Non-hydrogen atoms | 16,720 | 16,715 | |
| Protein residues | 16,175 | 16,175 | |
| Ligands | 545 | 540 | |
| B factors (Å2) | |||
| Protein | 154.41 | 244.56 | |
| Ligand | 133.45 | 169.20 | |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.004 | 0.004 | |
| Bond angles (°) | 1.032 | 1.031 | |
| Validation | |||
| MolProbity score | 1.53 (94th Percentile) | 1.56 (94th Percentile) | |
| Clashscore | 3.40 (97th Percentile) | 4.89 (94th Percentile) | |
| Poor rotamers (%) | 1.27 | 0.55 | |
| Ramachandran plot | |||
| Favored (%) | 95.61 | 95.61 | |
| Allowed (%) | 4.39 | 4.39 | |
| Disallowed (%) | 0.00 | 0.00 | |
