Abstract
The robust regenerative potential of skeletal muscle is imperative for the maintenance of tissue function across a host of potential insults including exercise, infection, and trauma. The highly coordinated action of multiple immune populations, especially macrophages, plays an indispensable role in guiding this reparative program. However, it remains unclear how skeletal muscle repair proceeds in a chronically inflamed setting, such as infection, where an active immune response is already engaged. To address this question, we used a cardiotoxin injury model to challenge the reparative potential of chronically infected muscle. Compared with regenerating naive skeletal muscle, infected skeletal muscle exhibited multiple indicators of delayed muscle repair including a divergent morphologic response to injury and dysregulated expression of myogenic regulatory factors. Further, using both flow cytometric and single-cell RNA sequencing approaches, we show that reduced macrophage heterogeneity due to delayed emergence of restorative subsets underlies dysfunctional tissue repair during chronic infection. Our findings highlight how the preexisting inflammatory environment within tissue alters reparative immunity and ultimately the quality of tissue regeneration.
Keywords: Infectious disease, Inflammation
Keywords: Bioinformatics, Macrophages, Skeletal muscle
Chronic infection alters the diverse macrophages that are required for skeletal muscle regeneration.
Introduction
As a highly regenerative tissue, skeletal muscle contains a major store of self-renewing myogenic precursor cells (satellite cells). Following injury, quiescent satellite cells activate, proliferate, fuse with other postmitotic activated satellite cells into multinucleated myotubes, and terminally differentiate (1). Each step of regeneration follows a preset program of gene expression that mimics its embryonic development involving myogenic master regulatory genes Pax7, Pax3, Myf5, Myod, and Myog (1–3). However, while satellite cells are the effectors of regeneration, inflammation and a highly coordinated immune program are indispensable for successful regeneration (4).
The requirement of a robust and organized immune response during skeletal muscle regeneration has been demonstrated in murine models of acute sterile injury, such as injection with the protein kinase C inhibitor cardiotoxin (CTX) (5–7). This program requires temporally distinct acute inflammatory and regenerative processes predominated by inflammatory and restorative macrophages, respectively (8). Inflammatory macrophages are responsible for the potentiation of an immune response, clearance of cellular debris, and activation of satellite cells, while restorative macrophages ameliorate inflammation, enhance extracellular matrix deposition, and promote satellite cell differentiation (7–9). Failure to efficiently transition from an inflammatory to a regenerative phase in a timely manner results in prolonged damage (6, 8, 9). Other cell types participate in this transitional phase. Specifically, skeletal muscle regulatory T cells (Tregs) rapidly accumulate during the transitional period and are critical in expediting this shift (6, 10). Also, satellite cells provide cues that facilitate transition of macrophage towards the proregenerative phenotypes (11).
Disruption of immune regulatory networks is a feature of many skeletal muscle diseases including inflammatory myopathies and muscular dystrophies (10, 12–14). In these pathogenic states of inflammation, decreased efficacy of muscle growth and repair can lead to increased muscle damage, fibrosis, stem cell exhaustion, and ultimately functional impairment (4, 15–17). Thus, it remains a pressing need to understand how chronically inflamed and nonsterile settings affect the orchestration of skeletal muscle immunity and the overall reparative program. Previously, we have shown that chronic infection with Toxoplasma gondii in skeletal muscle results in prolonged Th1 inflammation and muscle damage (18). Further, Tregs become pathogenic, actively inciting damage through promoting an inflammatory macrophage bias in the skeletal muscle, in contrast to promoting regenerative macrophages as previously described (6, 10, 18).
Whether the presence of long-standing infection-induced inflammation disrupts the ability of this tissue to respond to further insult remains unclear. In the present study, we used chronic infection with T. gondii as a model of skeletal muscle inflammation, to address if long-term inflammation impairs the progression of wound repair and the myogenic processes. We formally address how canonical immune- and tissue-reparative programs proceed following CTX-induced injury given the preexisting immune dysfunction present in chronically infected skeletal muscle. Intramuscular administration of CTX results in the depolarization of cell membranes, myofiber contraction, and tissue destruction (5). We find that chronically infected skeletal muscle undergoes distinct and divergent morphologic and molecular changes compared with naive skeletal muscle in response to toxin-induced injury, consistent with delayed repair and defective terminal differentiation of myofibers. Immunologically, infected-muscle macrophages are largely polarized to an inflammatory status and do not transition fully to restorative macrophages.
To date, the full phenotypic and functional heterogeneity of macrophage populations in skeletal muscle during injury or chronic infection has not been fully elucidated. Microarray analysis proved helpful to begin classifying the transcriptional changes macrophages undergo in response to injury induced by CTX (19). However, traditional genomic approaches are limited, as average gene readouts of bulk populations may mask biologically relevant macrophage subsets. We performed single-cell RNA sequencing of skeletal muscle macrophages and reveal for the first time to our knowledge the underlying cellular heterogeneity of macrophages during injury and infection. We further show that during CTX-induced injury in infected skeletal muscle, macrophages collectively retain an inflammatory signature similar to that during infection and share no overlap with differentially regulated genes identified in macrophages responding to CTX-induced injury alone. High-resolution unbiased single-cell clustering analysis reveals an absence of a critical population of reparative phagocytic and PPAR-signaling macrophages in CTX-injured infected muscle. Single-cell trajectory analysis demonstrates that the acquisition of reparative states is delayed in infected muscle compared with uninfected muscle responding to injury. We further show through flow cytometry that we can identify these populations and that they are also present and perturbed in the dystrophic muscle of mdx mice. Our results highlight a critical interaction between the existing inflammatory state of the tissue in altering macrophage heterogeneity and the tissue regenerative process in skeletal muscle.
Results
Chronic infection hinders the development of myofibers following acute sterile injury.
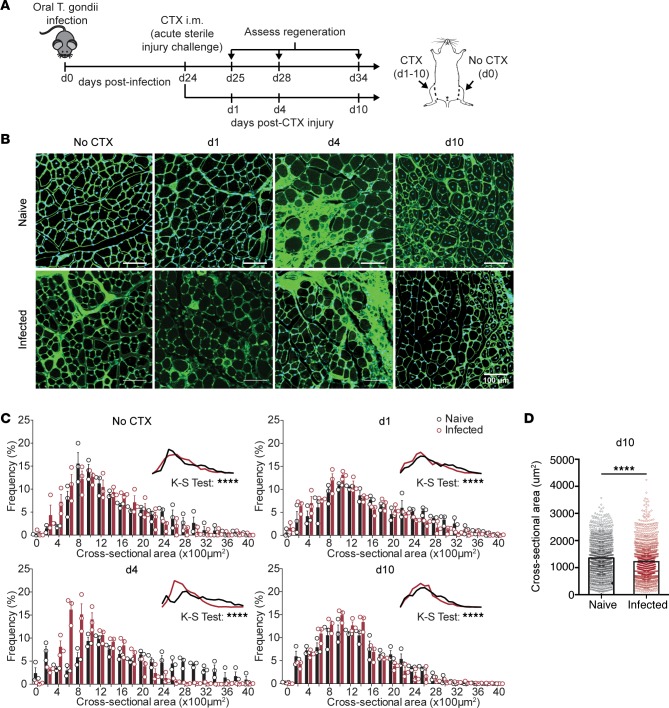
While the events mediating skeletal muscle repair have been well studied in uninfected tissue, little is known about how repair proceeds in the setting of chronic infection. To investigate whether chronic skeletal muscle infection alters the tissue’s ability to respond to subsequent injury, naive and chronically infected C57BL/6 mice were intramuscularly administered CTX 24 days after infection in one hindlimb (Figure 1A). We first addressed whether injury repair mechanisms were altered during chronic infection by monitoring and assessing morphological parameters of repair including percentage damaged area, myofiber cross-sectional area (CSA), and number of regenerating fibers. Muscle sections were prepared from mice 1, 4, and 10 days after CTX treatment and compared to the contralateral uninjured leg to monitor the morphological progression of degeneration and regeneration. As expected in the CTX-induced injury model, the greatest degree of damage was observed 4 days after CTX-induced injury (Figure 1B and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.121549DS1). In uninfected mice, this time point was characterized by the emergence of centrally nucleated, early regenerating fibers less than 400 μm in CSA, surrounded by a massive degree of inflammation (Figure 1C). In other studies, the inflammation present at 4 days after CTX has been shown to be dominated by restorative macrophages that facilitate the onset of the regenerative process (8, 9, 19). By 10 days after injury in uninfected muscle, the muscle architecture has returned histologically and larger regenerating fibers were abundant, as indicated by the number of centrally nucleated myofibers and myofiber CSA (Figure 1, B and D, and Supplemental Figure 1B).
Figure 1. Morphologic analysis of muscle regeneration following acute sterile injury in naive and chronically infected muscle.
(A) Experimental design for CTX-induced injury during chronic T. gondii infection. (B) Representative wheat-germ agglutinin (WGA, green) and DAPI (blue) staining of naive (top) and chronically infected skeletal muscle (bottom) during CTX-induced injury at days 0 (no CTX), 1, 4, and 10 after injury. Scale bars: 100 μm. (C) Fiber size frequency distribution of naive and chronically infected skeletal muscle at day 0 (no CTX), 1, 4, and 10 after CTX-induced injury (n = 3 mice/group). (D) Average muscle fiber cross-sectional area at 10 days after CTX-induced injury in naive and infected mice (n ≥ 1,000 measurements/group from n = 4 mice/group). Results are representative of 3 experiments of n = 3–4/group/experiment; error bars represent SEM (C and D). ****P < 0.0001 by Kolmogorov-Smirnov (K-S) test (C) or Student’s t test (D).
Consistent with our previous report, infected muscle exhibited a higher baseline level of damage before CTX-induced injury (Figure 1, B and C) (18). Four days after CTX, the extent of damage was equivalent to that of naive injured muscle (Figure 1, B and C). However, fibers from infected muscle remained a more homogeneous population of atrophied fibers (~600–1,000 μm CSA) with a reduced emergence of early regenerating fibers (<400 μm CSA), suggesting a delayed onset of the regenerative phase of repair. By 10 days after infection, the vast majority of myofibers were centrally nucleated and similar in number to the uninfected muscle (Supplemental Figure 1B). However, the CSA of the regenerating fibers was significantly smaller in infected muscle compared with naive muscle 10 days after CTX (Figure 1, C and D). Collectively, these findings indicate that chronic infection of the skeletal muscle leads to impairment in the morphologic characteristics of regenerating myofibers in response to toxin-induced damage.
Molecular progression of myogenesis is aberrant in infected mice.
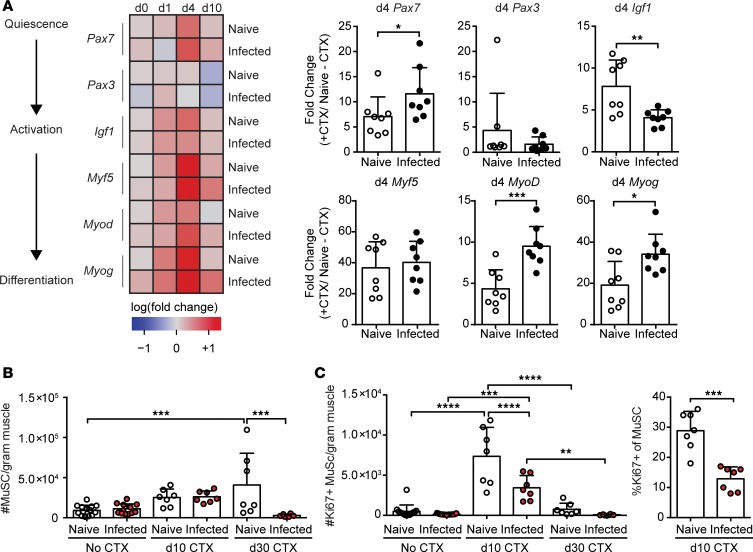
As the progression of myogenesis is governed by satellite cell activation and differentiation, we asked if there was an impediment of the reparative program at the molecular level. We first compared transcript levels of myogenic regulatory factors that are expressed at distinct times during the regenerative pathway from stem cell quiescence to terminal differentiation (Figure 2A). Day 1 following injury, the major satellite cell lineage–defining marker in postnatal muscle, Pax7, was quickly upregulated in naive muscle, but not in infected skeletal muscle (Figure 2A) (2). Pax3, a Pax7 paralog typically expressed during embryonic myogenesis, was upregulated in both naive and infected skeletal muscle (Figure 2A). However, despite its upregulation, Pax3-expressing satellite cells are known to comprise only a minor fraction of the overall stem cell population (2). Factors involved in early activation and proliferation (Igf1 and Myf5), early differentiation (Myod), and late differentiation (Myog) were expressed to similar degrees (Figure 2A) (1, 4, 20–23). By 4 days after injury, activation of pathways involved in all stages of repair, including Pax7, were highly upregulated in both naive and infected muscle (Figure 2A). As expected, these factors diminished in naive skeletal muscle by 10 days after CTX, indicating the successful resolution of the regenerative program in uninfected tissue (Figure 2A) (4). Notably, all myogenic regulatory factors, except Pax3, remained increased in infected skeletal muscle 10 days after CTX.
Figure 2. Molecular analysis of myogenic pathway following acute sterile injury in naive and chronically infected muscle.
(A) Whole muscle mRNA expression of muscle-repair associated genes, Pax7, Pax3, Igf1, Myf5, MyoD, and Myog relative to Gapdh relative to naive uninjured skeletal muscle throughout injury summarized as a heatmap (left) (n = 8 mice/group/day) and graphical summaries of gene expression at day 4 (right) (n = 8 mice/group). Data are cumulative of 2 independent experiments. Statistics performed on log-transformed values of relative expression. (B) Absolute number of muscle satellite cells (MuSCs: CD31–CD45–Sca1–CD106+) in naive and infected mice at 0, 10, and 30 days after infection. (C) Absolute number (right) and frequency of proliferating (Ki67+) MuSCs by flow cytometry in naive and infected muscle on days 0, 10, and 30 after CTX-induced injury. Error bars represent SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by 2-tailed Student’s t test (A), ANOVA with Tukey’s multiple-comparisons test (B and C, left), or Kruskal-Wallis with Dunnett’s multiple-comparisons test (C, right).
To directly investigate the proliferative capacity of satellite cells over the course of injury repair in uninfected and infected muscle, we compared the expression of the proliferation marker, Ki67, in satellite cells by flow cytometry directly ex vivo at 10 and 30 days after injury (24). At baseline, we find that there is no difference in the frequency or number of proliferating satellite cells between uninfected and infected muscle (Figure 2B). However, in response to injury, satellite cells from naive muscle exhibit a robust proliferative response 10 days after injury (Figure 2C). Notably, we observed a significant reduction in satellite cells at 30 days after injury in chronically infected muscle compared with uninfected muscle. This was accompanied by a significant reduction in satellite cell proliferation in infected muscle responding to injury (Figure 2C). Collectively, the impaired proliferation of satellite cells as well as the continued activation of the myogenic program and reduced myofiber size at 10 days after injury indicate that infection either delays the myogenic program or results in defective terminal differentiation of myofibers.
In situ transition from inflammatory to restorative macrophages is hindered in injured infected skeletal muscle.
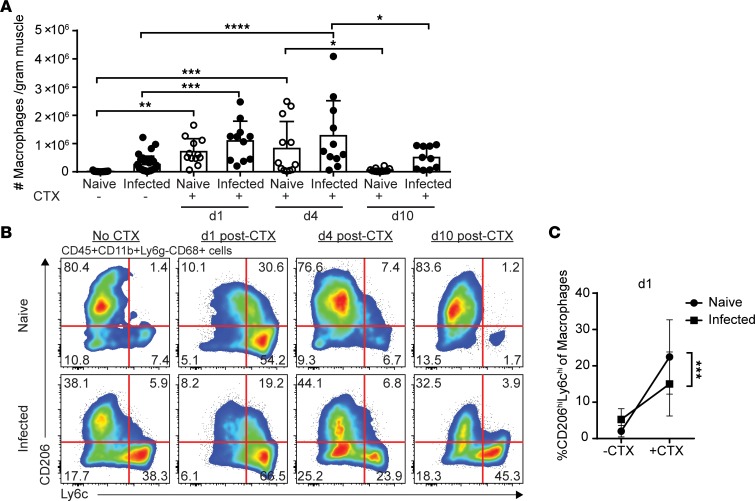
Macrophages are the major immune mediators of repair. Previous studies have shown that in response to acute sterile injury, inflammatory monocytes rapidly infiltrate the skeletal muscle and differentiate into inflammatory macrophages to facilitate clearance of debris, active myogenic precursors, and potentiate the inflammatory response (8, 19, 25). As repair progresses, these macrophages phenotypically switch in situ to become restorative macrophages, which support myogenesis and quell inflammation (8, 19). Consistent with this established paradigm, the total number of macrophages was highest at 1 and 4 days after injury but did not significantly differ between naive and infected mice at any time point (Figure 3A). We next addressed whether naive and infected-muscle macrophages transitioned between inflammatory and restorative subsets with the same kinetics and efficiency. Prior to injury, the majority of macrophages in uninfected muscle were of a tissue-resident/restorative phenotype (CD206hiLy6clo), purported to have more reparative and antiinflammatory properties, whereas infected muscle contained a sizeable population of inflammatory macrophages (Figure 3B and Supplemental Figure 2C) (18). One day after injury, the majority of macrophages in both naive and infected muscle were of an inflammatory phenotype (CD206loLy6chi) (Figure 3B and Supplemental Figure 2B). Interestingly, a small population of double-positive CD206hiLy6chi macrophages was also present at this time point, in agreement with observations of in situ differentiation of inflammatory macrophages toward the restorative phenotype (Figure 3B and Supplemental Figure 2; see refs. 8, 19). However, compared with naive muscle, CD206hiLy6chi transitional intermediate macrophages were significantly reduced in infected muscle (Figure 3, B and C). By days 4 and 10, the majority of macrophages had transitioned to restorative macrophages in uninfected muscle, while inflammatory macrophages still persisted within the infected skeletal muscle (Figure 3B). In agreement with a delayed transition of macrophages, we observed a dysregulation in the accumulation of Tregs and a sustained expression of Tbet throughout the response to CTX-induced injury in infected muscle compared with uninfected CTX-injured muscle (Supplemental Figure 3).
Figure 3. Transition toward restorative macrophages during regeneration is stunted in chronically infected muscle.
(A) Absolute number of total macrophages (CD45+CD11b+Ly6g–CD68+) per gram of skeletal muscle by flow cytometric analysis (n = 10–12 mice/group). (B) Representative flow plots and graphical summary of macrophage subsets (CD206hi/loLy6chi/lo) in naive and infected skeletal muscle at 0, 1, 4, and 10 days after CTX-induced injury (n = 10–12 mice/group). (C) Change in frequency of CD206hiLy6chi transitional macrophages 1 day after CTX-induced injury (n = 10–12 mice/group). Results are cumulative of 3 experiments of n = 3–4/group/experiment; error bars represent SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by ANOVA with Tukey’s multiple-comparisons test (A) or ANCOVA (C).
One explanation for the delay in the transition between inflammatory and restorative macrophages could be uncontrolled parasite reactivation due to the damage elicited by CTX to infected myofibers. To this end, we examined the parasite stage and overall parasite burden by measuring the expression of the tachyzoite-specific transcript Sag1 and the bradyzoite-specific transcripts Bag1 and Eno1 throughout the injury response, and normalized expression to uninjured infected muscle. We saw an increase in Sag1, Bag1, and Eno1 1 day after injury relative to uninjured infected muscle (Supplemental Figure 4A). Early upregulation of both tachyzoite- and bradyzoite-specific transcripts on day 1 may be an adaptive response of the parasite in preparation for egress and reinfection, as host cells are stressed by injury. Sag1 transcript levels returned to preinjury expression at day 4 and remained there at day 10. In contrast, Bag1 and Eno1 were decreased to below uninjured levels at day 4 and day 10 after injury (Supplemental Figure 4A). These data suggest that reactivation may transiently occur but is swiftly controlled by the host response. This was further supported by the lack of change in overall parasite burden in muscle throughout injury repair (Supplemental Figure 4B). Together, our findings reveal a decreased ability of macrophages to efficiently transition to a restorative phenotype following tissue injury in chronically infected muscle and is independent of changes in overall parasite burden.
Infection-induced transcriptional signature dominates in muscle macrophages during CTX-induced injury.
Macrophages exhibit remarkable plasticity in polarization among various tissue sites and under different stimulation conditions (26, 27). While traditional definitions of inflammatory and restorative macrophages are based on well-established markers in vitro, they do not capture the full diversity of macrophage phenotype, activation, and function in vivo and during injury repair in the muscle (19, 26, 28, 29). To gain a more in-depth understanding of the effects of infection on the response of muscle macrophages to acute CTX-induced injury, we assessed the transcriptional profile of muscle macrophages on both a global and single-cell level using single-cell RNA sequencing. To this end, we administered CTX to naive and infected mice as in Figure 1A. At day 4 after CTX-induced injury, we purified CD64+MerTK+ muscle macrophages from 3 treatment groups for single-cell RNA sequencing: (a) uninfected muscle with CTX-induced injury (CTX), (b) infected muscle without CTX-induced injury (infected), and (c) infected with CTX-induced injury (infected + CTX). Four days after injury was chosen, as this is the point at which the majority of muscle macrophages from uninfected CTX-injured muscle have shifted back to a restorative phenotype to begin the regenerative process, whereas infected CTX-injured muscle retains a mixed population of macrophages (Figure 3B and Supplemental Figure 2C; see refs. 5, 8, 19). For each treatment group, 1,630 cells or more were sequenced and analyzed.
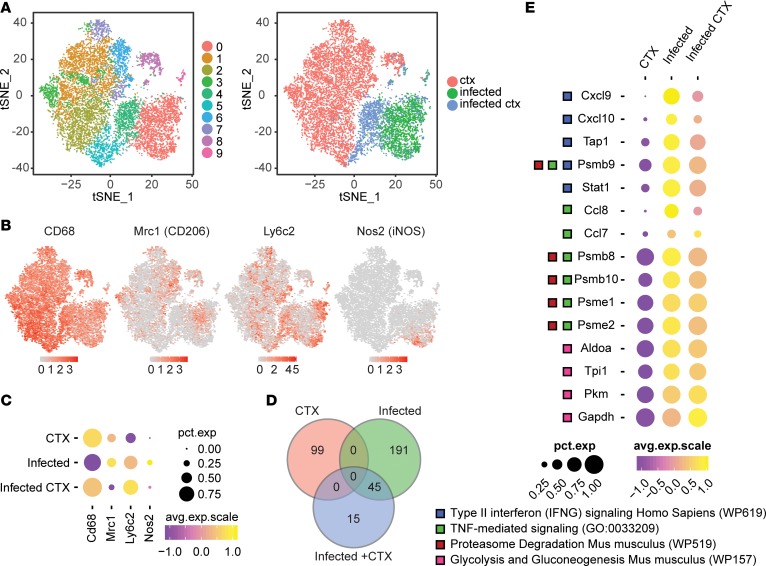
We first asked whether the macrophages from infected muscle responding to CTX-induced injury expressed a transcriptional profile similar to that of the reparative macrophage program from uninfected skeletal muscle responding to CTX. To compare the transcriptional profiles of macrophages from each treatment group relative to each other, we aggregated all events and performed unbiased cluster analysis based on statistically significant principal components identified by principal component analysis (PCA) using Seurat. Unsupervised cluster analysis identified 10 clusters, clearly visualized on a t-distributed stochastic neighbor-embedding (t-SNE) map (Figure 4A). When we annotated the identified clusters by their treatment group (Figure 4A), clusters 1–3 and 5–8 closely aligned with the CTX group, whereas clusters 0 and 9 closely aligned with the infected group. These 2 groups were nonoverlapping in high-dimensional space, suggesting significant differences in the principal components that defined these groups (Figure 4A). Interestingly, the infected + CTX group was composed of mainly clusters 4 and 5, which lie between the CTX and infected groups on the t-SNE plot, signifying shared similarities with both treatment groups (Figure 4A). To verify that our single-cell events were indeed macrophages, we confirmed the expression of the pan-macrophage marker Cd68 and plotted its scaled expression on the t-SNE map (Figure 4B). We also examined the expression of markers used previously by flow cytometry to define restorative and inflammatory macrophages, Mrc1 (CD206), Ly6c2 (Ly6c), and Nos2 (iNOS) (Figure 4B). Plots of the percentage of cells expressing of these markers in each treatment group, as well as the average expression of positively expressing cells, closely paralleled our flow cytometric protein-expression data (Figures 3B and 4C).
Figure 4. Transcriptional profiling of total macrophages during CTX-induced injury in uninfected and infected skeletal muscle.
Single-cell RNA sequencing was performed on macrophages isolated from uninfected CTX-injured (ctx), infected without CTX-injury (infected), and infected CTX-injured skeletal muscle. Data sets from each treatment group were aggregated for direct comparison. (A) t-SNE visualization of PCA and unsupervised cluster analysis performed on aggregated data set. Plot is annotated by bioinformatically identified clusters (left) and treatment group (right). (B) Scaled expression of Cd68, Mrc1, Ly6c2, and Nos2 overlaid on t-SNE plot of aggregated data set. (C) Dot plot representation of Cd68, Mrc1, Ly6c2, and Nos2 segregated by treatment group. Percentage expression of marker in each treatment group is represented by dot size. Average expression level in expressing cells is represented by color scale. (D) Venn diagram of differentially regulated genes identified in each treatment group. (E) GO and pathway enrichment analyses of 45 overlapping differentially regulated genes between infected and infected + CTX groups. Differentially regulated genes falling into selected significantly enriched terms and pathways are represented in the dot plot. Percentage expression of marker in each treatment group is represented by dot size. Average expression level in expressing cells is represented by color scale. Enriched terms and pathways were identified as significant at an adjusted P value ≤ 0.01 and FDR ≤ 0.05.
We next identified differentially expressed genes (DEGs) in the global macrophage population from each treatment group (CTX, infected, and infected + CTX). To do this, we used Seurat to cluster single macrophages based on their original treatment group. We then identified positively regulated genes from the averaged cumulative reads from each treatment group compared to all remaining cells. In this manner, we can computationally mimic bulk-RNA sequencing in order to globally assess changes in the overall macrophage compartment between experimental groups. Genes were considered differentially expressed if they were 2-fold positively increased with an adjusted P value of 0.05 or less and detected in at least 25% of cells from each comparison group. The top 15 DEGs from each treatment group were plotted on a heatmap (Supplemental Figure 5C). As expected, DEGs that were upregulated in the CTX group contained restorative macrophage-associated genes such as Fn1, Sepp1, and Cx3cr1, as well as genes related to phagocytosis such as Apoe and Cd63. Notably, we also observed a high upregulation of Igf1 in CTX macrophages (Supplemental Figure 5, A and B), which is consistent with the role of Igf1 in the restorative process (20, 22). We did not observe a significant upregulation of the promyogenic factors Il6, Il4, or Cxcl12 in any of the treatment groups (Supplemental Figure 5, A and B) (4). In contrast, proinflammatory markers such as Saa3, Lyz1, and Stat1 were upregulated in infected and infected + CTX groups. Comparison of complete lists of DEGs between the 3 treatment groups revealed a high degree of shared genes between the infected and the infected + CTX groups, but no shared genes with the CTX group (Figure 4D). Overlapping genes were significantly enriched for gene-ontology (GO) terms related to TNF-mediated signaling and pathways related to IFN-γ signaling, proteasome degradation, glycolysis, and gluconeogenesis (Figure 4E). These terms and pathways are associated with the classical activation of macrophages (29, 30). Because the clustered data set retains single-cell information, we can calculate the frequency of cells expressing a gene in a given treatment group. While the percentages of cells expressing the aforementioned genes were equivalent between infected and infected + CTX groups, the average level of expression was lower in the infected + CTX group (Figure 4E). Collectively, these data show that the global transcriptional profile of macrophages from chronically infected CTX-injured muscle did not resemble that of uninfected muscle responding to CTX-induced injury. Rather, they continued to exhibit a transcriptional signature adherent to proinflammatory function.
Identification and characterization of macrophage heterogeneity during CTX-induced injury.
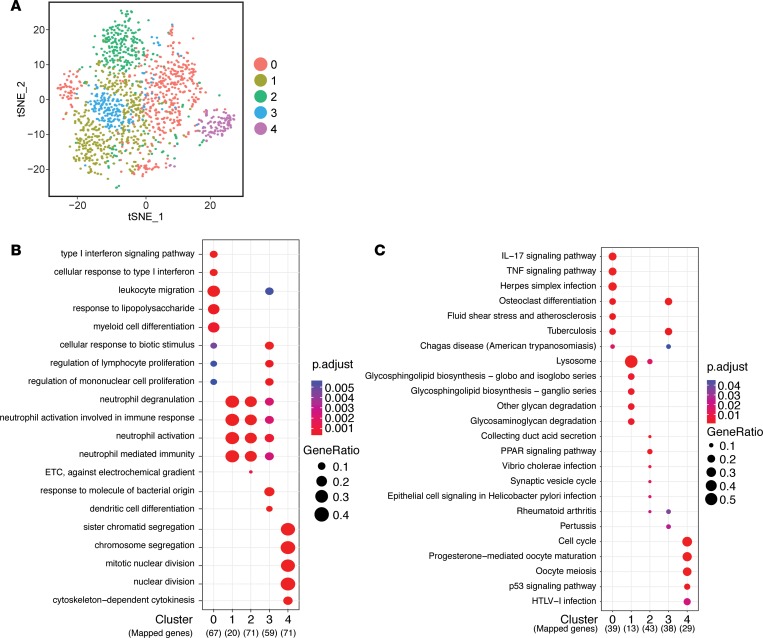
To better understand the changes occurring within the macrophage compartment during injury and infection, we increased the resolution of our analysis by focusing on the heterogeneity of macrophages that exist within each treatment group. In this manner, we were able to identify whether alterations in the composition of heterogeneous populations of macrophages occurred between treatment groups and characterized the functional relevance of these changes. Cluster analysis based on statistically significant principle components identified 5 distinct clusters of macrophages within the overall population of CTX skeletal muscle macrophages (Figure 5A). To identify whether these clusters represented unique populations, we compared lists of DEGs from each cluster to determine the degree of overlap between each cluster. DEG sets exhibited very little overlap with other clusters, with the exception of cluster 1 that shared genes with clusters 2 and 3 (Supplemental Figure 6A). The top 15 upregulated genes in each cluster were identified and visualized as a heatmap to confirm that the differentially regulated genes are highly specific for each cluster (Supplemental Figure 6B). To explore the potential role for each cluster of macrophages identified, we compared the biological themes represented in the lists of DEGs for each cluster by GO and pathway (KEGG) analyses using clusterProfiler. The top 5 GO terms or KEGG pathways for each cluster were plotted on the basis of an adjusted P value ≤ 0.01 and a false discovery rate (FDR) of ≤ 0.05 for identified annotations (Figure 5, B and C). While cluster 0 was highly upregulated in genes encoding canonical markers associated with restorative macrophages such as Mrc1 and Fn1, GO and KEGG analyses revealed enrichment for proinflammatory terms such as IFN, LPS, and TNF responses (Figure 5, B and C). Expression of both inflammatory and restorative macrophage–related transcripts is consistent with previous observations that macrophages transition from an inflammatory to a restorative phenotype between days 1 and 4. As expected based on the significant overlap in DEGs from cluster 1 with clusters 2 and 3, all three clusters contained a gene set enriched in GO terms related to neutrophil degranulation and function (Figure 5B). However, cluster 1 was also significantly enriched in KEGG pathways related to the lysosome and extracellular matrix degradation (Figure 5C). These results strongly indicate that cluster 1 represents a functional class of macrophages responsible for the breakdown of tissues to enable repair. In comparison, cluster 2 macrophages were highly enriched in pathways such as PPAR signaling (Figure 5C). PPARγ expression in macrophages has been previously shown to be a transcriptional regulator of reparative macrophages (31). Specifically, PPARγ controls the expression of growth differentiation factor 3 (GDF3), which promotes myoblast cell fusion during differentiation (31). Cluster 3 was similar to cluster 0 in that both clusters were highly enriched in GO terms related to classical activation such as cellular “response to biotic stimulus” and “response to molecule of bacterial origin” as well as KEGG pathways such as “Tuberculosis” and “Chagas Disease,” which require a robust inflammatory response from macrophages (Figure 5C). However, compared with cluster 0, cluster 3 did not highly express restorative macrophage–associated transcripts, rather macrophages from this cluster were highly upregulated in inflammatory transcripts, such as Il1b (Supplemental Figure 6B). Finally, DEGs in cluster 4 were significantly enriched in genes related to mitosis and cell cycle. Thus, this cluster represents a local highly proliferative macrophage population that populates the tissue during wound repair (8, 19, 32).
Figure 5. Identification and characterization of macrophage heterogeneity during tissue repair in uninfected muscle.
(A) t-SNE visualization of PCA and unsupervised cluster analysis from uninfected + CTX data set. (B and C) Top 5 terms identified by GO enrichment (B) and KEGG pathway (C) analyses for each cluster were plotted and compared between each cluster. Adjusted P value for each annotation is represented by color scale. Gene ratio is represented by dot size. Enriched terms and pathways were identified as significant at an adjusted P value ≤ 0.01 and FDR ≤ 0.05.
Identification and characterization of macrophage heterogeneity during infection.
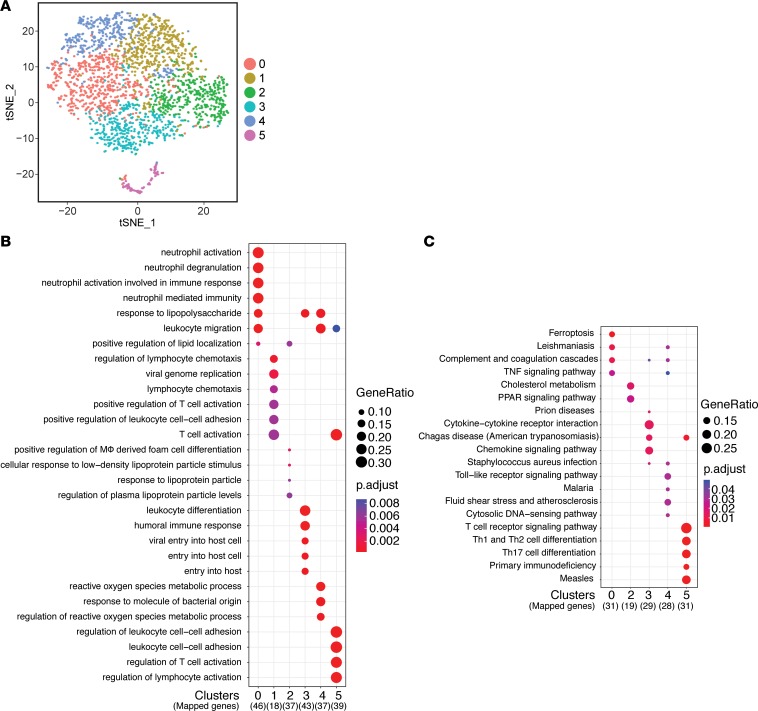
We next examined the heterogeneity of macrophages within chronically infected muscle. Cluster analysis of infected mice identified 6 distinct clusters and was visualized on a t-SNE map (Figure 6A). DEGs identified for each cluster were unique to their cluster, as each cluster had low overlap in the gene sets, with the exception of cluster 1 (Supplemental Figure 7A). The top 15 DEGs that comprised each cluster’s gene set were plotted as a heatmap, which confirmed the unique expression of these genes (Supplemental Figure 7B). To determine the functional relevance of each cluster, we again performed GO and KEGG analyses to compare biological themes between the clusters. Clusters 0, 3, and 4 represent an inflammatory set of macrophages that are enriched in terms and pathways related to classically activated macrophages and their proinflammatory function such as “response to lipopolysaccharide” and “leukocyte migration” (Figure 6B). However, while all 3 clusters shared an inflammatory signature, cluster 0 was specifically enriched for neutrophil-related pathways, cluster 1 was enriched for T cell activation, and cluster 3 was enriched for reactive oxygen pathways (Figure 6, B and C). These results suggest that the activating stimulus overlaps yet resulted in distinct functional roles for each cluster of macrophages. Cluster 1 highly expressed inflammatory monocyte-associated genes such as Ccr2 and Ly6c2 as well as genes annotated for T cell migration and activation, and therefore may represent a blood-derived inflammatory macrophage precursor population (Figure 6, B and C). In contrast to these clusters of inflammatory macrophages, cluster 2 macrophages were enriched in lipid metabolism pathways such as “PPAR signaling” ascribed to the reparative functions of macrophages (Figure 6C). Interestingly, cluster 5 expressed T cell–associated genes such as Cd3g, Cd8b1, and Gzmb, suggesting these are CD8+ T cells that were present in the preparation but were readily identified through bioinformatics in our analysis (Supplemental Figure 7B). This is validated by enrichment of terms and pathways associated with T cell activation and differentiation (Figure 6, B and C). These results further confirm our identification of macrophages within chronically infected muscle, but reveal a deeper understanding of the heterogeneity in both identity and function.
Figure 6. Identification and characterization of macrophage heterogeneity during chronic T. gondii skeletal muscle infection.
(A) t-SNE visualization of PCA and unsupervised cluster analysis from infected without CTX-induced injury data set. (B and C) Top 5 terms identified by GO enrichment (B) and KEGG pathway (C) analyses for each cluster were plotted and compared between each cluster. Adjusted P value for each annotation is represented by color scale. Gene ratio is represented by dot size. Enriched terms and pathways were identified as significant at an adjusted P value ≤ 0.01 and FDR ≤ 0.05.
Identification and characterization of macrophage heterogeneity during CTX-induced injury in infection.
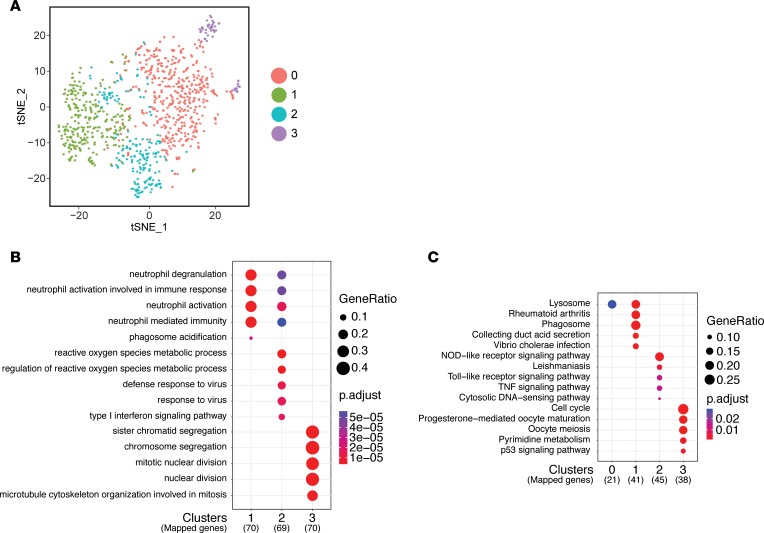
We next addressed the heterogeneity of macrophages in chronically infected muscle responding to CTX-induced injury. Cluster analysis of macrophages from CTX-injured infected muscle identified only 4 clusters (Figure 7A). Little overlap was detected in the DEG sets for each cluster (Supplemental Figure 8A). The distinct expression pattern for each cluster can be clearly visualized on a heatmap identifying the top 15 DEGs (Supplemental Figure 8B). KEGG analysis revealed that cluster 0 was highly enriched for genes involved in the lysosomal pathway, and shared an enriched gene set similar to CTX cluster 1 (Figure 5B and Figure 7, B and C). Clusters 1 and 2 were similar in the expression of inflammatory genes that converge on GO terms related to neutrophil degranulation and neutrophil activation (Figure 7B). However, distinguishing these clusters was the enrichment in lysosomal and phagosome pathways in cluster 1, versus enrichment of annotations related to reactive oxygen species and pathways involving TLR and TNF signaling in cluster 2, similar to infected cluster 3 (Figure 7, B and C). A relatively small population, cluster 3 is highly enriched in cell cycle related genes, and thus represents a locally proliferating population similar to CTX cluster 4 (Figure 7, B and C). While some clusters in the infected + CTX group share similarities with the CTX group, these clusters fail to fully recapitulate the full spectrum of macrophage heterogeneity observed in response to CTX-induced injury of uninfected muscle. These data suggest that a reduction in macrophage heterogeneity underlies the deficiency in a timely and robust regenerative response to CTX-induced injury during chronic infection.
Figure 7. Identification and characterization of macrophage heterogeneity during tissue repair in infected muscle.
(A) t-SNE visualization of PCA and unsupervised cluster analysis from infected + CTX data set. (B and C) Top 5 terms identified by GO enrichment (B) and KEGG pathway (C) analyses for each cluster were plotted and compared between each cluster. Adjusted P value for each annotation is represented by color scale. Gene ratio is represented by dot size. Enriched terms and pathways were identified as significant at an adjusted P value ≤ 0.01 and FDR ≤ 0.05.
Progression of macrophage states through wound repair is hindered during chronic infection.
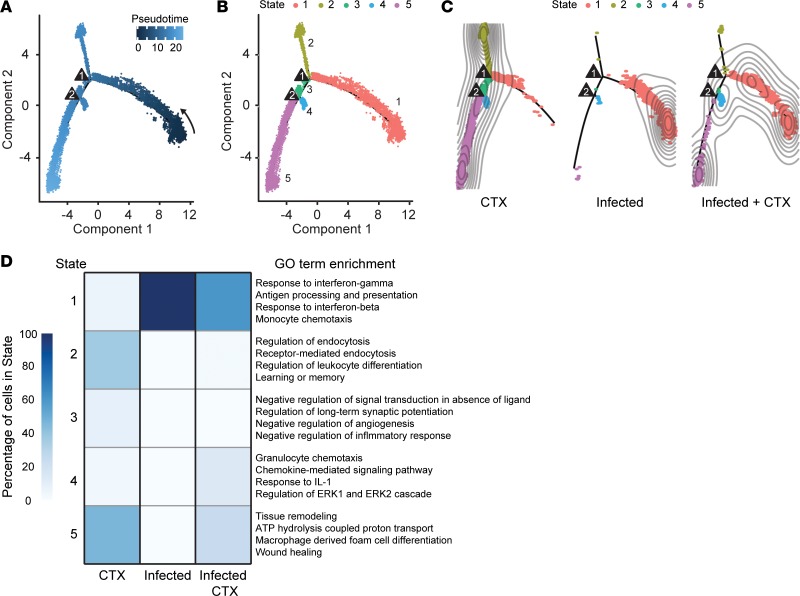
In response to CTX-induced injury, macrophages undergo dynamic transcriptional changes as they transition from one functional state to another (19). While macrophages from uninfected muscle responding to CTX-induced injury 4 days after injury are thought to predominantly adopt a reparative program, we show that a heterogeneous population exists representing macrophages from multiple stages of repair (Figure 5). Similarly, significant heterogeneity exists within macrophages of infected muscle (Figure 6). This heterogeneity underscores the transcriptional variability and transiency of macrophages as they proceed through a biological process such as wound repair. To gain further insight into how infection perturbs the transcriptional progression of macrophages during muscle repair, we used reverse graph embedding in Monocle 2 to identify endpoint/intermediate cell states and construct an unbiased single-cell trajectory through pseudotime, a measure of the progress a cell has made through a biological process (33, 34). To compare how cells from each treatment group track through pseudotime, the backbone for single-cell trajectories was generated using macrophages from infected, CTX-injured (day 4 after injury) uninfected, and CTX-injured infected muscle to map the full spectrum of differentiation states. Cells from each treatment group were ordered along pseudotime using an unbiased technique called dpFeature, which performs density cluster analysis (Figure 8A).
Figure 8. Macrophages from CTX-injured infected skeletal muscle are hindered in progression toward reparative macrophage populations through pseudotime.
(A) Single-cell trajectory through pseudotime constructed by reverse graph embedding of unbiased cluster analysis and dimensional reduction of single-cell transcriptomes from uninfected CTX-injured, infected, and infected CTX-injured macrophages. Triangles represent branch points. (B) Overlay of 5 identified differentiation states (Monocle states) on single-cell trajectory plot. (C) States parsed by treatment group on single-cell trajectory. Contours represent cell density. (D) Heatmap comparison of percentage of cells from each treatment group occupying individual states (left) and significantly enriched GO terms from differentially regulated genes of each state (right). Enriched terms were identified as significant at an adjusted P value ≤ 0.01 and FDR ≤ 0.05.
Single-cell trajectory analysis revealed 5 Monocle states (tree branches) with 2 bifurcation points indicative of fate decisions (Figure 8B). Parsing the Monocle states by treatment groups on the single-cell trajectory revealed that uninfected CTX-injured and infected macrophages occupy distinct Monocle states (Figure 8C). In contrast, infected CTX-injured muscle occupied multiple states overlapping with uninfected CTX-injured and infected macrophages, consistent the paradigm that macrophages from infected muscle are partly capable of responding to CTX-induced injury, and in agreement with our previous unbiased cluster analysis (Figures 7 and 8C). We next probed how macrophages from each treatment group were distributed among the different Monocle states. A small proportion of macrophages from uninfected CTX-injured muscle were found in state 1, which is characterized by enrichment of monocyte chemotaxis and inflammatory gene annotations (Figure 8D), suggestive of blood-derived inflammatory monocytes/macrophages. The majority of macrophages in this treatment group occupy states 2 and 5, which are associated GO annotations relaying phagocytic and tissue regenerative functions, respectively. States 3 and 4 are also represented in this group; however, they also only constitute minor populations. Macrophages in state 3 are enriched in regulatory function. Macrophages in state 4 are enriched in inflammatory amplification pathways, which may represent a subset of resident macrophages (given the positioning relative to regulatory/reparative macrophages) that first sense tissue damage and initiate the influx of monocytes/macrophages in state 1. Collectively, we show that a cross-sectional sampling of muscle macrophages 4 days after CTX-induced injury can be used to reconstruct a pseudotime single-cell trajectory that describes the transition from inflammatory to restorative macrophages.
During infection, the vast majority of macrophages at steady state were found in a continuum along the inflammatory state 1 (Figure 8, C and D). In response to CTX-induced injury, an increased portion of macrophages were also found in state 4 (inflammatory amplification), suggesting macrophages were appropriately sensing increased damage (Figure 8, C and D). The vast majority of macrophages continue to occupy state 1. However, some state 1 macrophages are shifted closer to branch point 1, similar to inflammatory monocytes/macrophages found in uninfected CTX-injured muscle (Figure 8, C and D). Strikingly, compared with uninfected CTX-injured muscle, no macrophages are found in state 2 (phagocytic) or 3 (regulatory) (Figure 8, C and D). Further, while an increased frequency of macrophages is found in state 5 (regeneration), the proportion compared with uninfected CTX-injured muscle is significantly less (Figure 8, C and D).
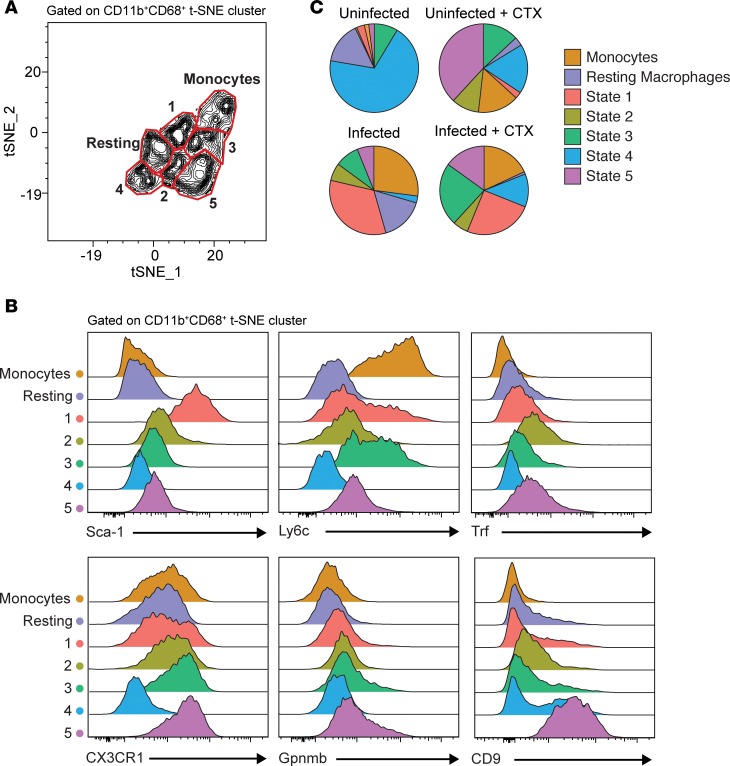
We next determined whether the heterogeneous macrophage states identified in Monocle could be monitored by flow cytometry. To do this we used highly upregulated genes from each of the Monocle states to design a panel of markers that could be used to distinguish these transcriptionally diverse populations. To interrogate our muscle leukocyte (CD45+) populations in an unbiased manner, we performed t-SNE analysis based CD11b, CD68, Sca-1, Ly6c, Trf, CX3CR1, Gpnmb, and CD9 to parse out CD11b+CD68+ macrophages expressing a unique constellation of state markers (Supplemental Figure 9, and Figure 9, A and B). From this analysis, we were able to identify 7 subsets of cells that corresponded to monocytes, resting macrophages, and Monocle states 1–5 based on expression of these markers (Figure 9, A and B). Finally, we verified that the proportion of macrophages that occupy each state is consistent with the frequencies calculated in our single-cell RNA sequencing analysis (Figures 8D and 9C, and Supplemental Figure 10C). We find that uninfected muscle responding to CTX-induced injury is able to assume a vast diversity of macrophage states 4 days after injury; they easily assume the regenerative states such as state 5 (Figure 9C). In contrast, the majority of macrophages in uninjured infected muscle are state 1 macrophages (Figure 9C). In response to CTX-induced injury, a large population of macrophages remains inflammatory, while few are found in reparative states like state 5 (Figure 9C). Collectively, these results validate our transcriptional analysis and demonstrate the ability of infection-induced chronic inflammation to restrict muscle macrophage mobilization toward CTX-associated restorative states (states 2–5) following acute muscle injury.
Figure 9. Flow cytometric validation of Monocle states identified by single-cell RNA sequencing.
t-SNE dimensional reduction analysis was performed on aggregated flow cytometric data sets of CD45+ cells from uninfected and infected muscle with or without CTX-induced injury (day 4). t-SNE clustering parameters were based on macrophage and Monocle state–distinguishing markers. (A) Contour plot and gating of density-rich, spatially distinct populations identified within the CD11b+CD68+ t-SNE cluster. Labeling and fine-tuning of subset gates (monocytes, resting macrophages, and Monocle states 1–5) were based on the phenotypic expression of corresponding Monocle state markers (n = aggregated data of all experimental groups, 7–8 samples/group). (B) Expression profile of Monocle state–distinguishing markers, Sca-1, Ly6c, Trf, CX3CR1, Gpnmb, and CD9 by flow cytometry of the individual gated subset populations (n = aggregated data of all experimental groups, 7–8 samples/group). (C) Distribution of Monocle states within total CD11b+CD68+ cells parsed by experimental group (n = 7–8 samples/group). Results are cumulative of 2 independent experiments of n = 3–4 samples/group/experiment.
Heterogeneous reparative macrophage populations are expanded in muscular dystrophy mice.
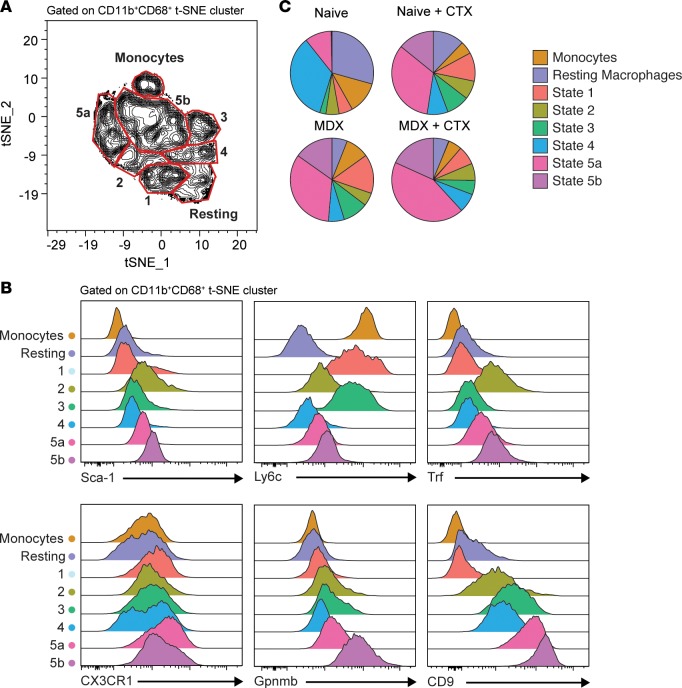
We next wanted to address the translatability of our findings to a different model of chronic skeletal muscle inflammation such as muscular dystrophy to better understand underlying disturbances in macrophage populations. To this end, we compared heterogeneous macrophage states identified previously in the mdx mouse model of muscular dystrophy to nondiseased (B10) muscle before and after CTX-induced injury (Supplemental Figure 10, A and B). Using our panel of markers, we again performed t-SNE on the total leukocyte population to serve as an unbiased approach to subsetting macrophages based on these markers (Figure 10A). Notably, while we could identify groups with profiles corresponding to monocytes, resting macrophages, and Monocle states 1–5, there was an additional group of macrophages that were similar to state 5, but with slightly lower expression of CD9 and Gpnmb (Figure 10, A and B, and Supplemental Figure 10D). For these reasons, we have denoted these populations as states 5a and 5b (Figure 10C). When comparing the distribution of macrophages in the skeletal muscle, we find that the mdx macrophages at steady state are very similar to uninfected muscle responding to CTX-induced injury (Figure 10B). This is consistent with the notion that there is constant injury and repair occurring in muscular dystrophy (10, 16, 35–37). In response to CTX-induced injury, mdx muscle is capable of expanding state 5a and 5b macrophages. Thus, in response to CTX-induced injury, macrophages in mdx muscle expand out restorative states. Collectively, our identification of unique states of functionally distinct macrophages are found within a variety of chronically inflamed and diseased skeletal muscle states.
Figure 10. Monitoring of macrophage heterogeneity in mdx mice at steady state and during injury repair.
t-SNE dimensional reduction analysis was performed on aggregated flow cytometric data sets of CD45+ cells from B10 and mdx muscle with or without CTX-induced injury (day 4). t-SNE clustering parameters were based on macrophage- and Monocle state–distinguishing markers. (A) Contour plot and gating of density-rich, spatially distinct populations identified within the CD11b+CD68+ t-SNE cluster. Labeling and fine-tuning of subset gates (monocytes, resting macrophages, and Monocle states 1–5) were based on the phenotypic expression of corresponding monocle state markers (n = aggregated data of all experimental groups, 10 samples/group). (B) Expression profile of Monocle state–distinguishing markers, Sca-1, Ly6c, Trf, CX3CR1, Gpnmb, and CD9 by flow cytometry of the individual gated subset populations (n = aggregated data of all experimental groups, 10 samples/group). (C) Distribution of Monocle states within total CD11b+CD68+ cells parsed by experimental group (n = 10 samples/group). Results are cumulative of 2 independent experiments of n = 5 samples/group/experiment.
Discussion
Skeletal muscle function is indispensable for many essential activities such as posture, movement, and voluntary breathing. However, its relatively superficial position makes it prone to insult. Thus, efficient tissue regeneration of this organ is critical to survival. Acute sterile injury models, such as CTX-induced injury, have afforded the ability to study the cellular, molecular, and immunologic responses that govern the normal regenerative process. These mechanisms serve as a benchmark to understand dysfunction in pathologic conditions. In this study, we highlight deficiencies in the muscle-specific response to CTX-induced injury in the context of chronic infection with T. gondii, which establishes a Th1-inflamed environment within the muscle. Understanding how reparative mechanisms are altered in this setting is of particular importance since injury rarely occurs in a sterile environment. Furthermore, devastating diseases such as inflammatory myopathies, muscular dystrophies, and age-related sarcopenia are similarly characterized by elements of chronic inflammation. Previously, identification of defects in tissue regeneration during chronic inflammation has proven challenging, as different fibers may be at varying stages of repair at any given time. By synchronizing injury through intramuscular injection of CTX, we demonstrate that the inability to coordinate heterogeneous macrophage populations, critical for repair, disrupts the myogenic program during chronic muscle infection. Furthermore, we extend our findings to the mdx model of muscular dystrophy and demonstrate that these unique states of functional macrophages are found during steady-state disease as well as in response to acute CTX-induced injury. Our findings highlight the deleterious impact of chronic inflammation on the ability to generate tissue reparative immune responses in muscle.
Previous studies elaborated the intricate morphologic and molecular changes that must occur in satellite cells to ensure a successful myogenic program. Our findings show that in chronic infection, the kinetics of early satellite cell proliferation and activation are delayed. This is further confirmed by a decrease in the detection of smaller regenerating myofibers. Similar delays in satellite cell responsiveness have also been characterized in dystrophy models (38, 39). Whether this is due to stem-cell exhaustion or simply the lack of proper extrinsic signals is unclear (38, 40, 41). While chronically infected muscle exhibits signs of ongoing damage and regeneration that typically accompanies stem cell exhaustion, the concurrent observation of increased expression of Pax7 at day 4 is incompatible with an exhaustion model (4). Our observation that satellite cells are significantly reduced in their proliferative potential 10 days after CTX-induced injury, which potentially results in a significant reduction in total satellite cells 30 days after CTX-induced injury, supports this idea. Together, these findings suggest that the intrinsic ability of the stem cell compartment to activate/proliferate is not totally impaired in chronic infection. One explanation for this could be that the impaired responsiveness may be a result of a paucity of environmental cues, as seen with the decreased whole tissue and macrophage expression of IGF-1 at day 4 after injury (20, 22).
A hallmark of regenerating myofibers is the presence of centrally located nuclei, which is clearly observed at 10 days after CTX-induced injury in uninfected muscle. While there are similar numbers of centrally nucleated fibers at 10 days after injury in chronically infected skeletal muscle, the comparatively smaller size of the regenerating myofibers and prolonged expression of myogenic regulatory factors (Myf5, Myod, and Myog) indicates a delay or deficiency in myofiber hypertrophy and maturation. The constellation of aberrant expression of myogenic regulatory factors, reduced satellite proliferation, and deviant morphologic changes suggest that infection-induced alterations to the tissue environment do not support proper growth of myofibers. Furthermore, these findings support the notion that effects of chronic infection and inflammation are primarily satellite cell extrinsic rather than intrinsic.
Macrophages are a major early source of IGF-1 during myogenesis (21, 42). Conditional knockout of IGF-1 in macrophages leads to dysfunction in satellite cell proliferation following injury, as well as a diminished shift to a CD206-expressing restorative macrophage phenotype (21). In our single-cell analysis, Igf1 expression is significantly upregulated in cluster 2 of CTX macrophages at 4 days after CTX-induced injury. Cluster 2 transcripts are also highly enriched for the “PPAR signaling” pathway, which has been shown to govern reparative functions of macrophages during myoblast differentiation (31). Notably, PPARγ signaling promotes production of GDF3, which regulates the fusion of regenerating fibers into multinucleated syncytial fibers (31). PPARγ expression has been shown to be upregulated in Ly6clo repair macrophages as early as day 2 after CTX-induced injury (31). We found during chronic infection a cluster of macrophages (cluster 2) enriched in the PPAR signaling pathway, as well as expressing canonical restorative macrophage markers such as Mrc1. This suggests that this macrophage population is capable of being generated in the setting of infection-induced chronic inflammation. However, a notable distinction is that Igf1 was not upregulated in this population like their PPAR signaling counterparts in uninfected CTX macrophages. Surprisingly, this cluster of macrophages is not present in infected muscle responding to CTX-induced injury 4 days after treatment. Thus, aberrant tissue regeneration during infection may be driven by interference with the timely emergence of PPARγ-expressing macrophages and sufficient accumulation at sites of local injury.
Another point of potential dysfunction is the relative absence of macrophages from certain Monocle states in CTX-injured infected muscle, as indicated in our single-cell trajectory analysis. Increased ApoE expression in macrophages has been shown to enhance phagocytosis of cellular debris during muscle regeneration (25). Diminished Apoe expression and macrophage-mediated debris clearance leads to stunted regeneration (25). We find that Apoe is enriched in 3 Monocle states (states 2, 3, and 5). As macrophages found in state 2 express the highest degree of Apoe and are enriched for genes involved in endocytosis, they likely constitute the highly phagocytic macrophages responsible for clearance of cellular debris. These macrophages are found in high abundance in uninfected CTX-injured muscle, but not in infected CTX-injured muscle. State 3 macrophages, which are enriched in genes annotated for immune regulatory function, are also relatively absent in infected CTX-injured muscle.
Following injury, damage is sensed, monocytes enter the tissue and phagocytose cellular debris, and macrophages undergo a metabolic and phenotypic shift toward a regenerative macrophage (19). This population of macrophages is sustained in the tissue by in situ proliferation (8, 19). Local proliferation of restorative macrophages instead of recruitment from the blood is thought to be a feature of the Th2 inflammatory environment (32). In this manner, restorative macrophage populations can accumulate in tissues without potentially injurious side effects of increased bone marrow activation or recruitment of other inflammatory cells from the blood (32). Despite the absence of a subset of PPAR signaling and highly phagocytic macrophages in infected + CTX-injured muscle, clusters of other repair-associated macrophages are present. Specifically, macrophages from the infected + CTX treatment group (clusters 0, 2, and 3) share overlap with 3 repair-associated clusters identified in the CTX group (clusters 1, 3, and 4). These clusters are characterized by enrichment of genes involved in regeneration, damage sensing, and cell cycle, respectively. Interestingly, proliferative and regenerative processes are thought to follow phagocytosis. Thus, macrophage populations conducive to muscle repair can accumulate in infected tissue following CTX-induced injury despite a reduced frequency of phagocytic macrophages. However, as evidenced by our single-cell trajectory analysis, the accumulation of these downstream populations is reduced and is correlated with the inability to generate other upstream populations such as phagocytic macrophages in a timely fashion. Together, these observations support the idea that injured infected muscle exhibits impairment in the progression of macrophages through defined regenerative transcriptional states.
When assessing global gene expression in macrophages from the 3 treatment groups, uninfected + CTX, infected, and infected + CTX, no shared genes were detected between uninfected and infected muscles or uninfected and infected muscles responding to CTX-induced injury. However, as discussed above, at single-cell resolution, identification of macrophage heterogeneity in each group reveals shared clusters of macrophages between CTX-injured groups in both uninfected and infected, thus showing the power that single-cell RNA sequencing provides. These clusters of similar macrophages are likely masked in the global assessment due to the predominance of inflammatory macrophages in infected + CTX muscle. Both infected and infected + CTX muscle share significant overlap in differentially regulated genes adherent to an inflammatory transcriptional program and promote glycolytic metabolism, both known to be upregulated in classically activated macrophages. Single-cell analysis reveals that these inflammatory macrophages are not uniform in their transcriptional signature, but rather relay distinct functions associated with phagocytosis, T cell activation, degranulation, or reactive oxygen species metabolism. The retention of these inflammatory macrophage populations in infected muscle following CTX-induced injury suggest their programming is refractory to exogenous or intrinsic signals that drive restorative macrophage polarization. The IFN-γ–rich environment of the bone marrow during T. gondii infection was previously shown to alter monocyte precursor gene expression (43). Accordingly, conditioning in this setting potentially gives rise to refractory inflammatory macrophages in the tissues during chronic infection. Likewise, conditioning in the tissue itself may reinforce an inflammatory trait.
An ongoing presence of inflammatory macrophages refractory to signals leading to an acquisition of restorative macrophage function may lead to niche competition detrimental to tissue regeneration. While injury-responsive inflammatory monocytes readily fill the niche of uninfected muscle, niche availability in infected muscle may be limited due to the presence of these refractory inflammatory macrophages required for immune surveillance. As such, recently emigrated inflammatory monocytes that participate in acute inflammation, phagocytosis, and ultimately differentiation into restorative macrophages following injury may represent a much smaller proportion of cells in infected muscle.
Another factor that may direct the quality of wound repair and the transition of inflammatory macrophages to restorative macrophages is Treg activity. We previously showed that Tregs acquire pathogenic function during chronic infection with T. gondii (18). In comparison with CTX-injured uninfected muscle, where Tregs accumulate with the same kinetics at this critical transitional period and highly expressed RORγt (6), we find Treg accumulation is delayed and Tbet expression is sustained in injured-infected muscle. The delayed onset of accumulation is associated with a concomitant decrease in the frequency of transitioning macrophages 1 day after CTX-induced injury. A defect in the regenerative capacity of aged muscle has previously been attributed to a lack of Treg accumulation following tissue injury (44). Given the wide transcriptional changes that occur in Treg plasticity, discrepancies in the way Tregs coordinate macrophages may also be influenced by inflammatory adaptations, such as the presence of IFN-γ (45–48).
While satellite cells are the engine of muscle repair, the inflammatory response to injury is essential to the efficacy of repair (6, 16, 44, 49). A comprehensive understanding of how these newly identified heterogeneous populations of macrophages collaborate to facilitate efficacious repair in homeostatic conditions and how this heterogeneity is perturbed during chronic inflammation is necessary to identify novel therapeutic pathways for a wide range of myopathies. Using single-cell RNA sequencing and unbiased cluster analysis, we show for the first time to our knowledge the full transcriptional heterogeneity of macrophages responding to injury at the single-cell level both in uninfected and infected tissues. Flow cytometric and single-cell trajectory analyses of skeletal muscle macrophages reveal a defect in the ability of chronically infected muscle to assemble the full repertoire of macrophage heterogeneity necessary to facilitate timely and efficacious repair. Our findings highlight how the preexisting inflammatory environment within tissue alters reparative immunity and ultimately the quality of tissue regeneration.
Methods
Mice.
Female C57BL/6 mice were obtained from Taconic Farms. C57BL/10 and C57BL/10ScSn-Dmdmdx/J (mdx) mice were obtained from Jackson Laboratories.
Infection with T.
gondii. Brain cysts for infection were prepared from chronically infected mice (≥30 days after infection) as previously described (18). Eight- to 12-week-old female mice were infected by oral gavage with 5 ME49 cysts.
Muscle injections.
For acute CTX-induced injury, anesthetized mice were injected with 30 μl/muscle of Naja pallida CTX (0.03 mg/ml) (Latoxan) in one or more hindlimb muscles. Uninjected contralateral hindlimbs were used as uninjured controls.
qRT-PCR of muscle differentiation factors.
RNA from murine muscle was isolated by TRIzol extraction and treated with DNase (Ambion). Purified RNA was converted to cDNA (iScript, BD Biosciences) and assayed for muscle differentiation factors Pax7 (forward: 5′-GACTCGGCTTCCTCCATCTC-3′, reverse: 5′-AGTAGGCTTGTCCCGTTTCC-3′), Pax3 (forward: 5′-ACTACCCAGACAATTTACACCAGG-3′, reverse: 5′-AATGAGATGGTTGAAAGCCATCAG-3′), IGF1 (forward: 5′-GTGTGGACGAGGGGCTTTTACTTC-3′, reverse: 5′-GCTTCAGTGGGGCACAGTACATCTC-3′), Myf5 (forward: 5′-GAACAGGTGGAGAACTATTA-3′, reverse: 5′-GCACATGCATTTGATACATCAG-3′), MyoD (forward: 5′-GAGCGCATCTCCACAGACAG-3′, reverse: 5′-AAATCGCATTGGGGTTTGAG-3′) and Myog (forward: 5′-CCAGTACATTGAGCGCCTAC-3′, reverse: 5′-ACCGAACTCCAGTGCATTGC-3′) by qPCR (iTaq Universal SYBR Green supermix, BD Biosciences). Ct values were normalized to the housekeeping gene Gapdh (forward: 5′-CCCACTCTTCCACCTTCGATG-3′, reverse: 5′-GTCCACCACCCTGTTGCTGTAG-3′).
Quantification of parasite burden.
DNA and RNA were extracted from infected murine muscle with the DNeasy Blood and Tissue kit (Qiagen) or TRIzol (Life Technologies), respectively. Total parasite burden was quantified by PCR amplification of the T. gondii–specific gene B1 from DNA isolated from target tissues (forward: 5′-TCCCCTCTGCTGGCGAAAAGT-3′, reverse: 5′-AGCGTTCGTGGTCAACTATCGATTG-3′). Ct values obtained from experimental samples were compared to a standard curve based on B1 amplification of known T. gondii DNA concentrations. Tachyzoite parasite burden was quantified by qRT-PCR of Bag1 (forward: 5′-GGGATGTACCAAGCATCCTG-3′, reverse: AGGGTAGTACGCCAGAGCAA-3′). Bradyzoite parasite burden was quantified by qRT-PCR of Sag1 (forward: 5′-ATCGCCTGAGAAGCATCACTG-3’, reverse: 5′-CGAAAATGGAAACGTGACTGG-3′), and Eno1 (forward: 5′-GGTATTGATATGCTTATGGTGGAG-3′, reverse: 5′-GCGATGTATTTGTATAGTGGTAGG-3′). TgActin (forward: 5′-ATGTATGTCGCTATCCAGGCCGTT-3′, reverse: 5′-TGATCTTCATGGTGGAAGGAGCCA-3′) was used as the housekeeping gene.
Muscle histopathology.
Muscle histology was performed as previously described (18, 50). Briefly, tibialis anterior muscles were perfusion fixed in 4% paraformaldehyde (Sigma-Aldrich) and isolated. Muscle samples were cryopreserved in 30% sucrose and subsequently frozen in OCT (Sakura) and cryosectioned at 20 μm. Sections were stained with wheat germ agglutinin (WGA) conjugated with Alexa Fluor 488 (1:100, Thermo Fisher Scientific) and DAPI (Sigma-Aldrich). Images were acquired with a Zeiss Axio Imager Z1 microscope. Image analysis was performed using FIJI software. Percentage damaged area was determined by binarization of the WGA-stained area as a percentage of total muscle area. CSA was quantified by segmentation analysis and subsequent measurement. The number of regenerating myofibers was quantified by enumerating the number of centrally nucleated myofibers normalized to total muscle area.
Isolation of tissue lymphocytes and satellite cells and flow cytometry.
Cells from the spleen and skeletal muscle were isolated as previously described (18). Satellite cells were isolated as previously described (24). Single-cell suspensions were stained with HBSS containing LIVE/DEAD Fixable Aqua dead cell stain (Life Technologies) and extracellular antibody stains. Cells were then fixed in 4% paraformaldehyde (EMD Millipore) and permeabilized in 0.5% saponin (Sigma-Aldrich), or fix/permeabilized in combination (Intracellular Fixation and Permeabilization Buffer Set, eBioscience). Afterwards, cells were rinsed and stained with eBioscience Permeabilization Buffer containing intracellular antibody stains. Samples were washed and resuspended in flow cytometry buffer (PBS, 1% bovine serum albumin [Sigma-Aldrich], 2 mM EDTA [Life Technologies]) for acquisition. Absolute numbers were calculated using CountBright absolute counting beads (Life Technologies).
Antibodies.
The following fluorescently conjugated antibodies used were obtained from BD Biosciences, BioLegend, or eBioscience/Thermo Fisher Scientific: TCRβ-APC-Cy7 (clone H57-597), CD4-PE-Cy7 (RM4-5), CD11b-BV605 (M1/70), Ly6G-PECF94 (1A8), CD45-V450 (30-F11), RORγt-PerCP-Cy5.5 (Q31-378), Foxp3-FITC (FJK-16s), Tbet-ef660 (eBio4B10), Ly6C-PerCp-Cy5.5 (HK1.4), CD68-PE-Cy7 (FA-11), Gpnmb-APC (CTSREVL), streptavidin-PE CD206-APC (C068C2), CD64-Pacific Blue (x54-5/7.1), MerTK-PE (2b10c42), CD45-APC (30-F11), CD31-APC (MEC13.3), Zombie Green fixable live/dead stain (Biolegend), Sca1-Pacific Blue (D7), CD106-Biotin [429(MVCAM.A)], CD9-Biotin (MZ3), CD206-FITC (C068C2), and streptavidin-BV650. Flow cytometry data were acquired using a BD LSRFortessa and analyzed using FlowJo (Tree Star).
Sort-purification of muscle macrophages.
Single-cell suspensions of skeletal muscle lymphocytes were obtained as above. Cells were surface stained for CD45, CD64, and MerTK. Macrophage populations were bulk purified using a BD FACSAria I into complete media.
Single-cell sequencing and quality control (10× Genomics Pipeline).
Cells were spun and resuspended in 1× PBS for counting on the Luna Automated Cell Counter (Logos Biosytems) using a 1:1 (v/v) dilution with 0.4% trypan blue. Cells were further diluted to approximately 200 cells/μl in 1× PBS and recounted as before. Approximately 2,000–5,000 cells were then added onto the 10× Genomics Chromium platform chip in combination with 3′ transcriptome capture reagents and single-cell capture was performed according to the manufacturer’s recommendations. Following capture, cDNA was prepared on a Bio-Rad T1000 overnight and double-stranded DNA libraries were prepared. After a final Ampure XP (Beckman Coulter) bead cleanup, libraries were quantified using Qubit fluorescence and assessed using the Advanced Analytics Fragment Analyzer using standard reagents. Absolute concentration of libraries was achieved using the Kapa Biosystems Universal qPCR protocol according to the manufacturer. Following qPCR, libraries were denatured and diluted to 8 pM for loading onto a HiSeq 2500 in RAPID mode with 26 × 98 paired-end-run parameters.
Fastq files were generated using the 10× Genomics CellRanger v2.02 analysis package. Briefly, cellranger mkfastq demultiplexed the raw bcl files into fastq files. Then, each sample was analyzed using the cellranger count function, aligning and quantifying each sample against the prebuilt 10× references for the mm10 mouse genome. The resulting alignments were reviewed for quality using 10× Cell Loupe Browser software.
Bioinformatics software and analysis.
Single-cell RNA data obtained from the 10× Genomics platform were analyzed using R 3.4.3 and RStudio. R packages used include Seurat (51), Monocle 2 (33, 34), UpSet (52), and clusterProfiler (53). Only cells where at least 200 genes were detected were included in downstream analysis. Outliers with ≥4,100 genes were excluded from analysis. Filtered cells were log-normalized and scaled for downstream analysis. PCA, clustering, t-SNE, identification of differentially regulated genes, and generation of heatmaps was performed on Seurat (details and walkthrough can be found at http://satijalab.org/seurat/). Pseudotime analysis was performed using Monocle 2. A single-cell trajectory and Monocle states were constructed by reverse graph embedding using cells sequenced from all treatment groups (http://cole-trapnell-lab.github.io/monocle-release/). Differentially regulated cells in each Monocle state were identified using Seurat. Upset Venn diagram plots were generated using lists of differentially regulated genes identified by Seurat. GO and KEGG analyses were performed on differentially regulated gene lists and compared across cell clusters using clusterProfiler.
Data availability.
The data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE113111).
Statistics.
Two-tailed Student’s t test was used to asses 2 independent groups. To compare the distribution of 2 independent variables, a Kolmogorov-Smirnov test was used. For comparison of multiple groups on parametric parameters, 1-way ANOVA was used with Tukey’s correction for multiple comparisons. For comparison of multiple groups on nonparametric parameters, a Kruskal-Wallis test was used with Dunnett’s correction for multiple comparisons. Analysis of covariance (ANCOVA) was used when appropriate to control for covariates such as pretest scores when assessing differences in dependent variables. Statistics were performed on log-transformed values for qPCR analysis. DEGs in RNA sequencing experiments were defined as genes with a fold change ≥2 at P ≤ 0.05. Enriched terms and pathways were identified as significant at an adjusted P value of ≤ 0.01 and FDR ≤ 0.05. Statistics for bioinformatics analysis was performed within the respective R package. All other statistics were generated using GraphPad Prism v6.0c.
Study approval.
All mice were bred and maintained under pathogen-free conditions at an American Association for the Accreditation of Laboratory Animal Care–accredited animal facility at the University at Buffalo under an animal study proposal approved by the University at Buffalo IACUC committee. All procedures involving mice were reviewed and approved by the IACUC at the University at Buffalo.
Author contributions
RMJ and EAW conceived and designed the studies. RMJ and JW performed the experiments and acquired data. RMJ and EAW analyzed the data and wrote the manuscript.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the NIH under award number R21AI128284 (to EAW). We thank the Confocal Microscope and Flow Cytometry Core Facility at the Jacobs School of Medicine, the University at Buffalo Genomics and Bioinformatics Core, and the Roswell Park Comprehensive Cancer Center Department of Flow and Image Cytometry for their technical assistance; and members of the Wohlfert lab and Dawn Bowdish and Chelsie Armbruster for helpful discussion and critical reading of this manuscript.
Version 1. 09/20/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interests exists.
Reference information: JCI Insight. 2018;3(18):e121549. https://doi.org/10.1172/jci.insight.121549.
References
- 1.Buckingham M, Rigby PW. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 2014;28(3):225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 2.Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomès D, Tajbakhsh S. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005;19(12):1426–1431. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ten Broek RW, Grefte S, Von den Hoff JW. Regulatory factors and cell populations involved in skeletal muscle regeneration. J Cell Physiol. 2010;224(1):7–16. doi: 10.1002/jcp.22127. [DOI] [PubMed] [Google Scholar]
- 4.Tidball JG. Regulation of muscle growth and regeneration by the immune system. Nat Rev Immunol. 2017;17(3):165–178. doi: 10.1038/nri.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardy D, et al. Comparative study of injury models for studying muscle regeneration in mice. PLoS ONE. 2016;11(1):e0147198. doi: 10.1371/journal.pone.0147198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burzyn D, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155(6):1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng B, Wehling-Henricks M, Villalta SA, Wang Y, Tidball JG. IL-10 triggers changes in macrophage phenotype that promote muscle growth and regeneration. J Immunol. 2012;189(7):3669–3680. doi: 10.4049/jimmunol.1103180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnold L, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204(5):1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Melton DW, Porter L, Sarwar ZU, McManus LM, Shireman PK. Altered macrophage phenotype transition impairs skeletal muscle regeneration. Am J Pathol. 2014;184(4):1167–1184. doi: 10.1016/j.ajpath.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villalta SA, et al. Regulatory T cells suppress muscle inflammation and injury in muscular dystrophy. Sci Transl Med. 2014;6(258):258ra142. doi: 10.1126/scitranslmed.3009925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patsalos A, et al. In situ macrophage phenotypic transition is affected by altered cellular composition prior to acute sterile muscle injury. J Physiol (Lond) 2017;595(17):5815–5842. doi: 10.1113/JP274361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figarella-Branger D, Civatte M, Bartoli C, Pellissier JF. Cytokines, chemokines, and cell adhesion molecules in inflammatory myopathies. Muscle Nerve. 2003;28(6):659–682. doi: 10.1002/mus.10462. [DOI] [PubMed] [Google Scholar]
- 13.Waschbisch A, Schwab N, Ruck T, Stenner MP, Wiendl H. FOXP3+ T regulatory cells in idiopathic inflammatory myopathies. J Neuroimmunol. 2010;225(1–2):137–142. doi: 10.1016/j.jneuroim.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt J, Rakocevic G, Raju R, Dalakas MC. Upregulated inducible co-stimulator (ICOS) and ICOS-ligand in inclusion body myositis muscle: significance for CD8+ T cell cytotoxicity. Brain. 2004;127(Pt 5):1182–1190. doi: 10.1093/brain/awh148. [DOI] [PubMed] [Google Scholar]
- 15.Mann CJ, et al. Aberrant repair and fibrosis development in skeletal muscle. Skelet Muscle. 2011;1(1):21. doi: 10.1186/2044-5040-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosurgi L, Manfredi AA, Rovere-Querini P. Macrophages in injured skeletal muscle: a perpetuum mobile causing and limiting fibrosis, prompting or restricting resolution and regeneration. Front Immunol. 2011;2:62. doi: 10.3389/fimmu.2011.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mojumdar K, et al. Inflammatory monocytes promote progression of Duchenne muscular dystrophy and can be therapeutically targeted via CCR2. EMBO Mol Med. 2014;6(11):1476–1492. doi: 10.15252/emmm.201403967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin RM, Blair SJ, Warunek J, Heffner RR, Blader IJ, Wohlfert EA. Regulatory T cells promote myositis and muscle damage in Toxoplasma gondii infection. J Immunol. 2017;198(1):352–362. doi: 10.4049/jimmunol.1600914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varga T, et al. Highly dynamic transcriptional signature of distinct macrophage subsets during sterile inflammation, resolution, and tissue repair. J Immunol. 2016;196(11):4771–4782. doi: 10.4049/jimmunol.1502490. [DOI] [PubMed] [Google Scholar]
- 20.Barton ER, DeMeo J, Lei H. The insulin-like growth factor (IGF)-I E-peptides are required for isoform-specific gene expression and muscle hypertrophy after local IGF-I production. J Appl Physiol. 2010;108(5):1069–1076. doi: 10.1152/japplphysiol.01308.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tonkin J, et al. Monocyte/macrophage-derived IGF-1 orchestrates murine skeletal muscle regeneration and modulates autocrine polarization. Mol Ther. 2015;23(7):1189–1200. doi: 10.1038/mt.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang SY, Goldspink G. Different roles of the IGF-I Ec peptide (MGF) and mature IGF-I in myoblast proliferation and differentiation. FEBS Lett. 2002;522(1–3):156–160. doi: 10.1016/s0014-5793(02)02918-6. [DOI] [PubMed] [Google Scholar]
- 23.Wolf JR, Hirschhorn RR, Steiner SM. Growth factor responsiveness: role of MyoD and myogenin. Exp Cell Res. 1992;202(1):105–112. doi: 10.1016/0014-4827(92)90409-2. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Cheung TH, Charville GW, Rando TA. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat Protoc. 2015;10(10):1612–1624. doi: 10.1038/nprot.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnold L, et al. CX3CR1 deficiency promotes muscle repair and regeneration by enhancing macrophage ApoE production. Nat Commun. 2015;6:8972. doi: 10.1038/ncomms9972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rigamonti E, Zordan P, Sciorati C, Rovere-Querini P, Brunelli S. Macrophage plasticity in skeletal muscle repair. Biomed Res Int. 2014;2014:560629. doi: 10.1155/2014/560629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray PJ, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varga T, et al. Macrophage PPARγ, a lipid activated transcription factor controls the growth factor GDF3 and skeletal muscle regeneration. Immunity. 2016;45(5):1038–1051. doi: 10.1016/j.immuni.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins SJ, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332(6035):1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu X, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14(10):979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison J, Lu QL, Pastoret C, Partridge T, Bou-Gharios G. T-cell-dependent fibrosis in the mdx dystrophic mouse. Lab Invest. 2000;80(6):881–891. doi: 10.1038/labinvest.3780092. [DOI] [PubMed] [Google Scholar]
- 36.Vidal B, et al. Fibrinogen drives dystrophic muscle fibrosis via a TGFbeta/alternative macrophage activation pathway. Genes Dev. 2008;22(13):1747–1752. doi: 10.1101/gad.465908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villalta SA, Deng B, Rinaldi C, Wehling-Henricks M, Tidball JG. IFN-γ promotes muscle damage in the mdx mouse model of Duchenne muscular dystrophy by suppressing M2 macrophage activation and inhibiting muscle cell proliferation. J Immunol. 2011;187(10):5419–5428. doi: 10.4049/jimmunol.1101267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blau HM, Webster C, Pavlath GK. Defective myoblasts identified in Duchenne muscular dystrophy. Proc Natl Acad Sci USA. 1983;80(15):4856–4860. doi: 10.1073/pnas.80.15.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang NC, Chevalier FP, Rudnicki MA. Satellite cells in muscular dystrophy - lost in polarity. Trends Mol Med. 2016;22(6):479–496. doi: 10.1016/j.molmed.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sacco A, et al. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143(7):1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boldrin L, Zammit PS, Morgan JE. Satellite cells from dystrophic muscle retain regenerative capacity. Stem Cell Res. 2015;14(1):20–29. doi: 10.1016/j.scr.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu H, Huang D, Saederup N, Charo IF, Ransohoff RM, Zhou L. Macrophages recruited via CCR2 produce insulin-like growth factor-1 to repair acute skeletal muscle injury. FASEB J. 2011;25(1):358–369. doi: 10.1096/fj.10-171579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Askenase MH, et al. Bone-marrow-resident NK cells prime monocytes for regulatory function during infection. Immunity. 2015;42(6):1130–1142. doi: 10.1016/j.immuni.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuswanto W, et al. Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin-33-dependent accumulation of regulatory T cells. Immunity. 2016;44(2):355–367. doi: 10.1016/j.immuni.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wohlfert EA, et al. GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J Clin Invest. 2011;121(11):4503–4515. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panduro M, Benoist C, Mathis D. Treg cells limit IFN-γ production to control macrophage accrual and phenotype during skeletal muscle regeneration. Proc Natl Acad Sci USA. 2018;115(11):E2585–E2593. doi: 10.1073/pnas.1800618115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hall AO, et al. The cytokines interleukin 27 and interferon-γ promote distinct Treg cell populations required to limit infection-induced pathology. Immunity. 2012;37(3):511–523. doi: 10.1016/j.immuni.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sefik E, et al. Mucosal immunology. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science. 2015;349(6251):993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castiglioni A, et al. FOXP3+ T cells recruited to sites of sterile skeletal muscle injury regulate the fate of satellite cells and guide effective tissue regeneration. PLoS One. 2015;10(6):e0128094. doi: 10.1371/journal.pone.0128094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin RM, Warunek J, Wohlfert EA. Therapeutic administration of IL-10 and amphiregulin alleviates chronic skeletal muscle inflammation and damage induced by infection therapeutic administration of IL-10 and amphiregulin alleviates chronic skeletal muscle inflammation and damage induced by infection. Immunohorizons. 2018;2(5):142–154. doi: 10.4049/immunohorizons.1800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33(18):2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE113111).