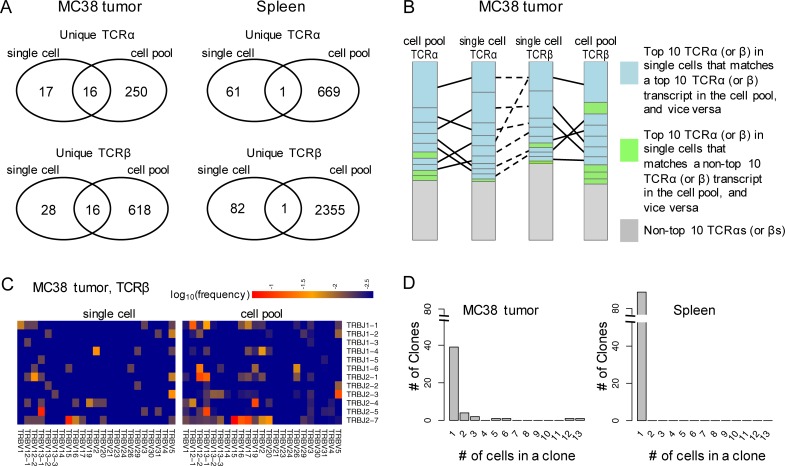
Fig 1. TCR profiling using the full length single cell RNA-Seq data.
A) Unique TCRα and TCRβ sequences detected in the CD8+ T cells from the MC38 tumor and the spleen by single cell RNA-Seq, in comparison to the RNA-Seq of the aliquot of cell pools used for the single cell capture; B) Consistency of TCRα and TCRβ sequences detected in the MC38 tumor and in the spleen by RNA-Seq of single cells and the aliquot of cell pools used for the single cell capture. Each block represents one TCRα (or TCRβ) sequence with the height of the block proportional to its clonal abundance (based on the cell counts) in the single cell data or the transcript abundance (based on the read counts) in the cell pool data, respectively. Matches between the top 10 abundant TCRα (or TCRβ) sequences detected by either approach are connected by solid lines and the associated sequences are colored blue. Sequences detected by both approaches but ranked within the top 10 by only one are colored green. In the single cell dataset, the TCRα and TCRβ sequences observed in the same cell are connected with a dashed line. One TCRβ can be connected to two TCRαs (or vice versa) in cells with a biallelic TCRα (or TCRβ) locus. C) Usage of the TRBV and TRBJ genes in the MC38 tumor infiltrating T cells, measured in the single cells (left panel) and in the corresponding cell pool (right panel). The union of the TRBV (and TRBJ) genes detected in the two approaches are presented. D) Distribution of TCR clones in the MC38 tumor and the spleen detected by the single cell RNA-Seq. A clone refers to the unique pairing of TCRα and TCRβ sequences.