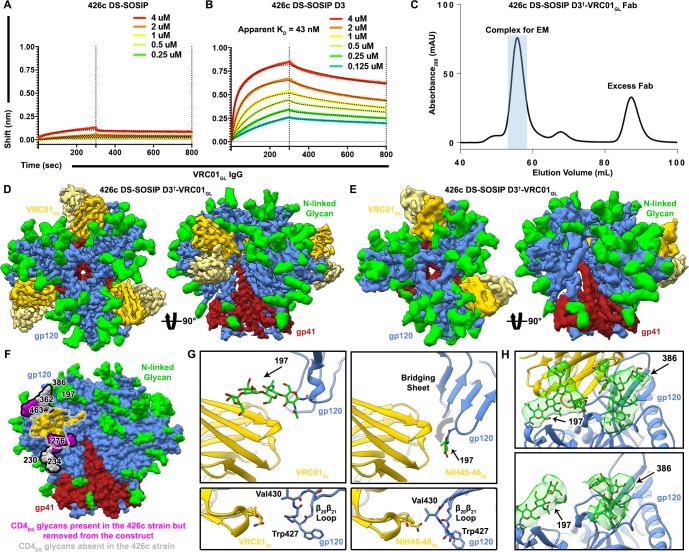
Figure 1. Structural characterization of the 426c DS-SOSIP D3†-VRC01GL complex.
(A–B) BLI binding data of immobilized VRC01GL IgGs binding to WT 426c DS-SOSIP (A) or 426c DS-SOSIP D3 trimers. The concentrations of 426c DS-SOSIP trimers injected are indicated on each panel. Fit curves are colored as black dotted lines. A KD could not be determined in (A) due to the weak responses observed. The vertical dotted lines indicate the transition between association and dissociation phases. (C) Size-exclusion chromatogram of the purified 426c DS-SOSIP D3†-VRC01GL complex used for cryoEM structure determination. The pooled fractions used for cryoEM are highlighted in light blue. (D) Two orthogonal views of the 3.8 Å cryoEM reconstruction sharpened with a B-factor of −250 Å2 whereas the glycan density is shown unsharpened. (E) Two orthogonal views of the asymmetric 4.8 Å reconstruction with two bound Fabs. (F) Surface representation of the 426c SOSIP trimer highlighting differences in glycosylation compared to the BG505 SOSIP. Glycans not present in 426c are colored light-gray and outlined. Glycans present in the 426c strain but removed by mutation from the 426c DS-SOSIP D3† construct are colored magenta and outlined. The gp120 surface buried at the interface with VRC01GL is indicated as a dotted outline and is colored yellow. (G) Comparison of the gp120 bridging sheet conformation when VRC01GL-class Fabs are bound to either 426c DS-SOSIP D3† trimer (Top-left) or a previously solved 426c gp120 core lacking selected NLGSs, such as the Asn276 NLGS (PDB: 5IGX) (Top-right). Comparisons of β20β21 loop conformations of each complex are shown below corresponding top panels. (H) Comparison of glycan density and position between VRC01GL-bound and VRC01GL-free protomers in the asymmetric cryoEM reconstruction shown in (E). (Top) Asn197 and Asn386 glycan density is stronger for protomers bound to VRC01GL Fab than for the gp120 protomer not bound to VRC01GL (Bottom). In panels D-H, gp120 protomers are shown in blue, gp41 in red, N-linked glycans in green and VRC01GL in dark and light yellow for the heavy and light chains, respectively.
Figure 1—figure supplement 1. Multiple sequence alignment of analyzed HIV-1 426c constructs.
Figure 1—figure supplement 2. Multiple sequence alignment of analyzed antibody and Fab constructs.
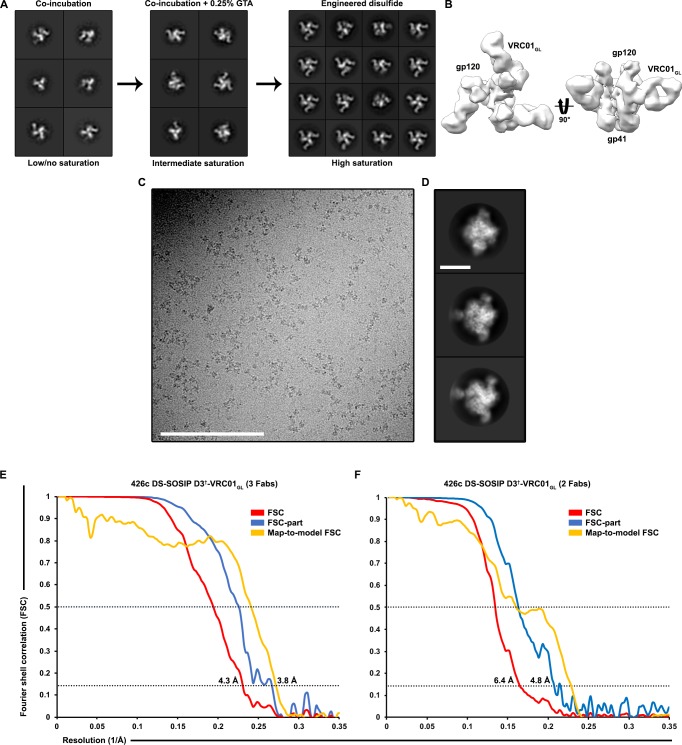
Figure 1—figure supplement 3. Structural characterization of the 426c DS-SOSIP D3†-VRC01GL complex.
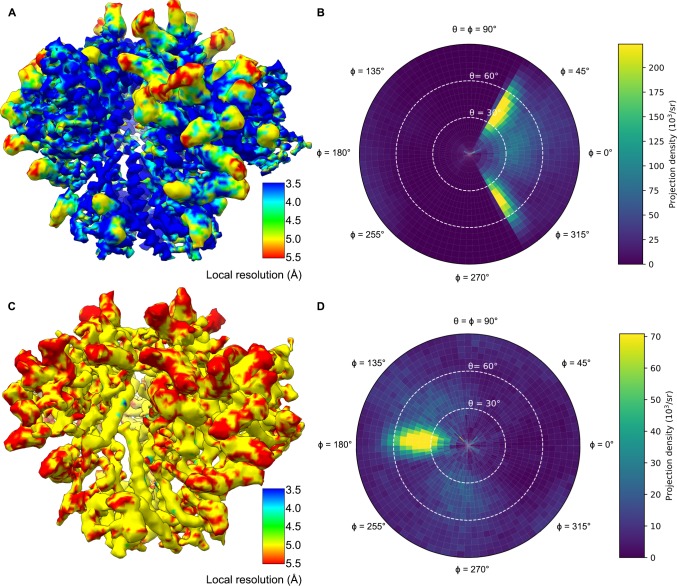
Figure 1—figure supplement 4. Validation of the 426c DS-SOSIP D3†-VRC01GL cryoEM reconstructions.
Figure 1—figure supplement 5. Comparison of gp120 interface contacts between VRC01GL and VRC01MAT.
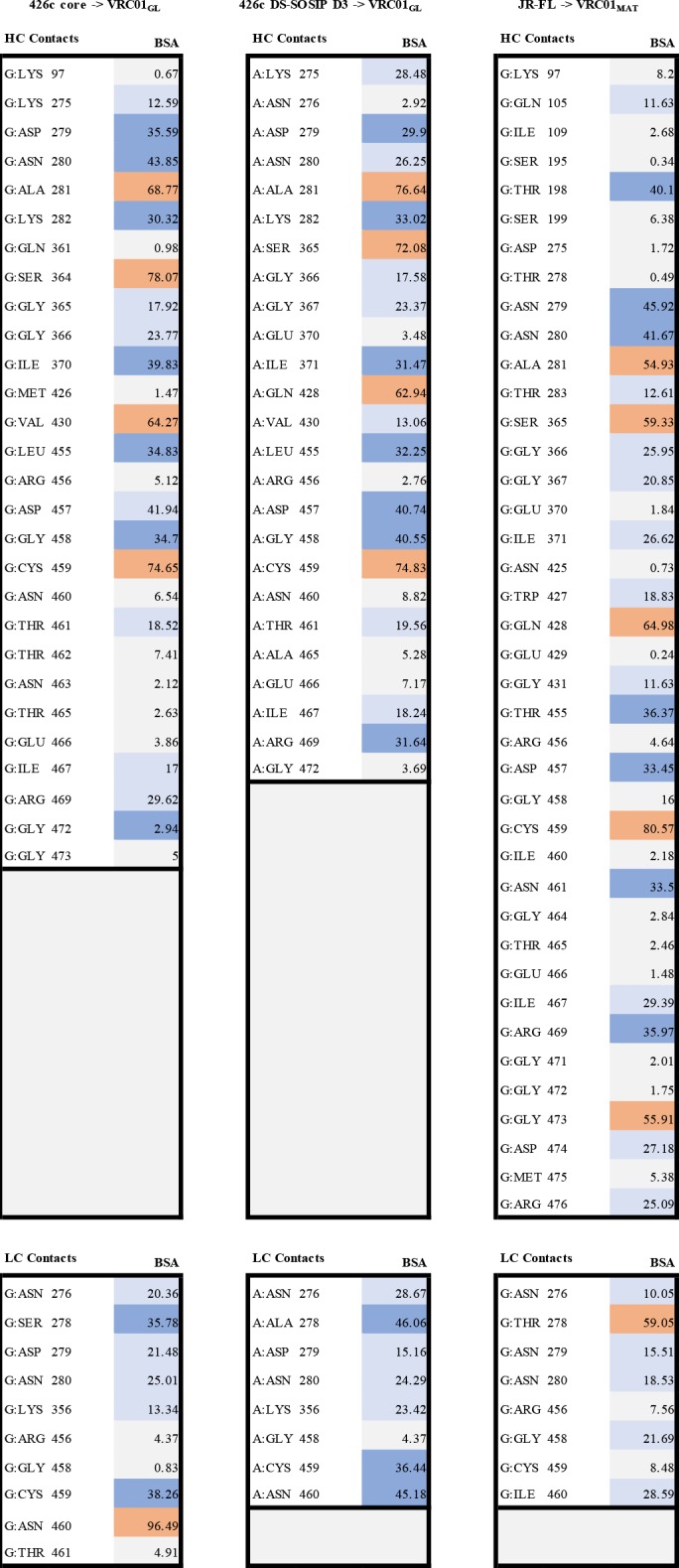
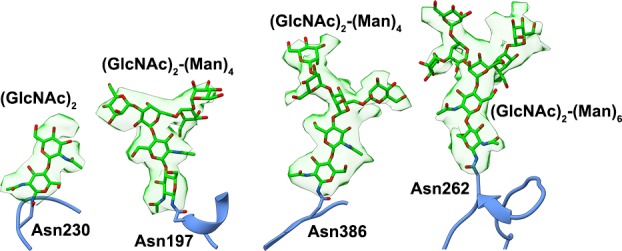
Figure 1—figure supplement 6. Example of glycans resolved in the 426c DS-SOSIP D3†-VRC01GL structure.