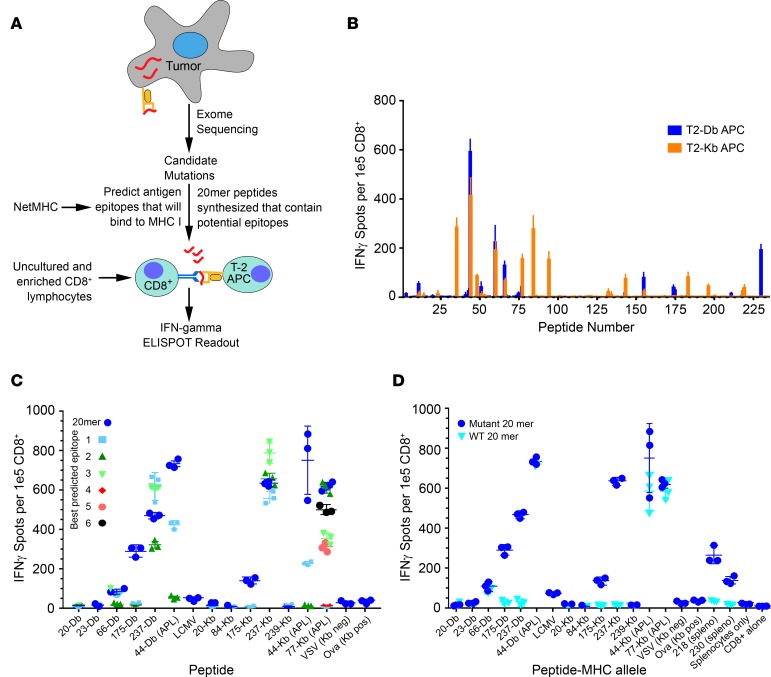
Figure 1. Neoantigen prediction pipeline and estimation of immunogenicity.
(A) Tumor neoepitope identification pipeline from whole-exome sequencing (WES) to in vivo verification. (B) C57BL6J mice were immunized with 20-mer peptides corresponding to mutant Panc02 peptides identified in Supplemental Table 4. Isolated CD8+ T cells were stimulated with T2-Db or Kb APCs pulsed with cognate peptides on an IFN-γ capture plate and resulting spots were counted (ELISPOT assay, see Methods). (C) Mice were immunized with pooled 20-mer peptides, and isolated CD8+ cells were tested for reactivity to minimal epitopes that were predicted from NetMHC algorithm (Supplemental Table 3). Altered peptide ligands (APLs) are indicated in the graph. The peptide number is followed by the MHC-restricted allele (Db or Kb) on the x axis. Symbols represent a single mouse together with mean ± SEM (n = 3 mice per group). Negative controls were VSV and LCMV peptides (see Methods). (D) Mice were immunized with pooled 20-mer peptides corresponding to the mutant neoepitopes and APLs, and isolated CD8+ T cells were analyzed for cross-reactivity to the wild-type 20-mer.