Figure 1.
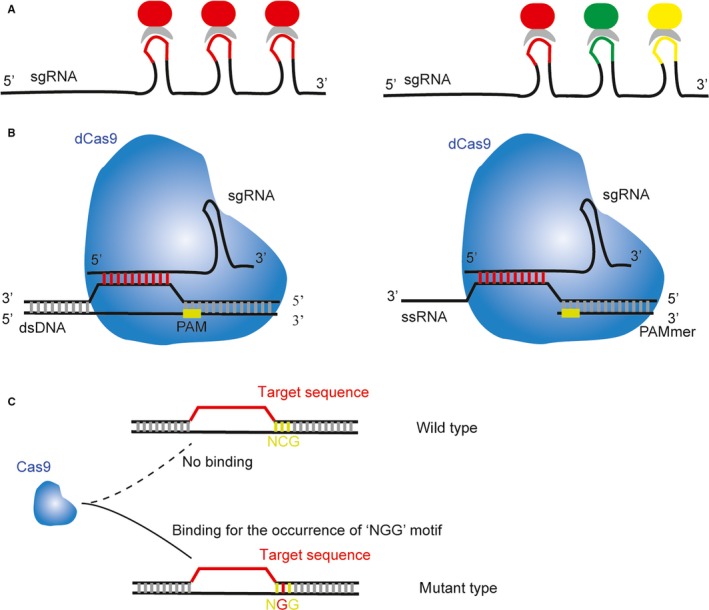
The applications of CRISPR/Cas system in molecular detection. A, Extension of the 3′ end of sgRNA to add extra hairpins. The RNA‐binding proteins (RBPs) are grey. The fluorescent proteins (FPs) are highlighted in red, green, and yellow. The left‐hand panel shows signal amplification effects of the addition of an extra hairpin structure for increasing the number of FPs for labelling the RBP. The right‐hand panel illustrates application of multicolour labels via different hairpin structures and a corresponding RBP and FP. B, Customization of dCas9/sgRNA for binding to DNA or RNA. The left‐hand panel shows that recognition of the double‐stranded DNA (dsDNA) by the sgRNA is determined by the 20 specific nucleotides of the sgRNA and by the PAM motif on the complementary strand close to the target region. The right‐hand panel shows that the artificial PAMmer composed of a deoxyribonucleotide is designed to assist sgRNA in recognition of a single‐stranded RNA (ssRNA), because there is no PAM motif in the ssRNA. C, One of the results of a single‐nucleotide variant (SNV) is creation of a novel PAM because of the appearance of the “NGG” motif, which makes the binding between Cas9 and the mutant DNA possible
