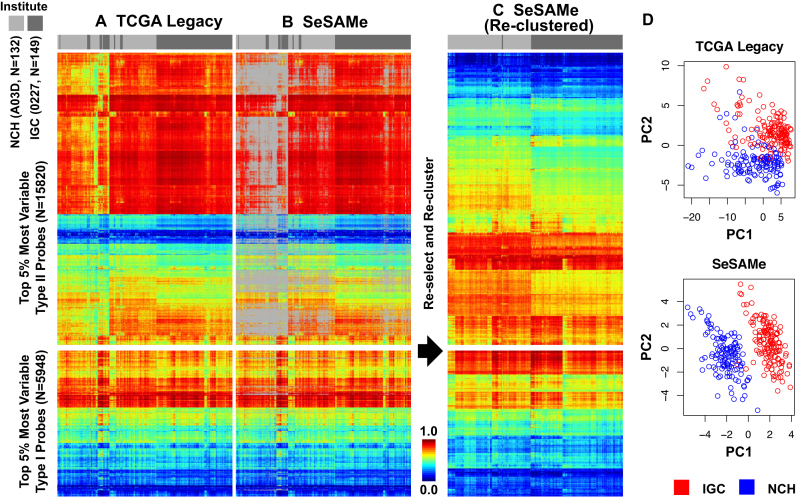
Figure 7.
SeSAMe preprocessing improves clustering of TCGA cell line replicates driven by small biological differences associated with two different institutes (IGC and NCH with barcodes 0227 and A03D, respectively) performing initial independent expansion of the same cell line (See text). (A) Clustering heatmap showing incomplete separation of 0227 and A03D replicates (columns) in existing TCGA Legacy DNA methylation 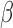 values, based on top variable probes (rows); (B) SeSAMe effectively masks spurious intermediate methylation visible in A; (C) Clustering after SeSAMe masking, with top variable probes (rows) re-selected based on SeSAMe’s masking and re-clustered. Samples are now better clustered by whether they are 0227 or A03D; (D) PCA analysis showing the first two principal components (PC1 and PC2) in TCGA Legacy data and SeSAMe remasked data.
values, based on top variable probes (rows); (B) SeSAMe effectively masks spurious intermediate methylation visible in A; (C) Clustering after SeSAMe masking, with top variable probes (rows) re-selected based on SeSAMe’s masking and re-clustered. Samples are now better clustered by whether they are 0227 or A03D; (D) PCA analysis showing the first two principal components (PC1 and PC2) in TCGA Legacy data and SeSAMe remasked data.