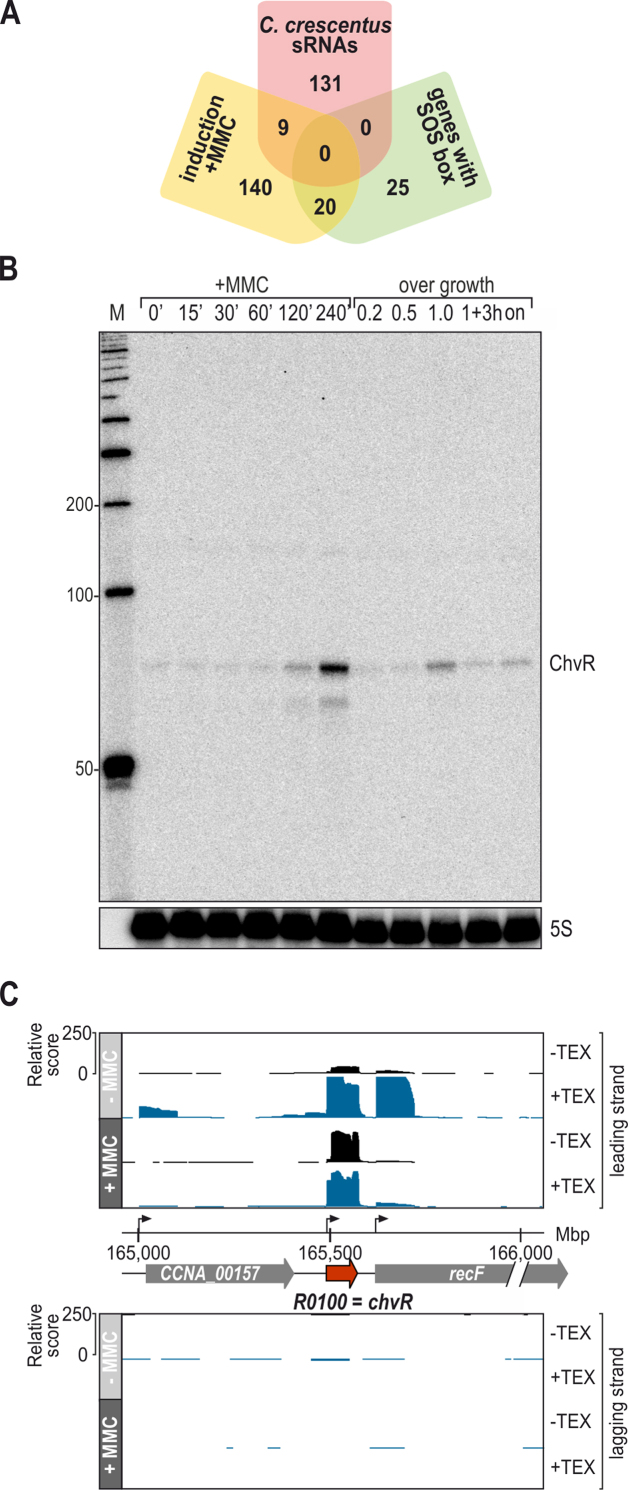
Figure 1.
Transcriptome analysis of C. crescentus in response to DNA damage. (A) Diagram summarizing the results of the dRNA-seq experiment. RNA was collected from C. crescentus grown in PYE prior to (OD660 of 0.4) or 240 min after treatment with the DNA-damaging agent MMC. Under these conditions, 169 genes were induced >3-fold, including 9 sRNAs. No conserved SOS box was identified in proximity to transcriptional start sites of sRNAs. (B) Expression of ChvR sRNA in wild-type C. crescentus. Cells were grown in PYE, and RNA samples were collected prior to (OD660 of 0.4) and at indicated time-points after MMC treatment, or at different time-points over growth (OD660 of 0.2, 0.5, 1.0, 3h after cells had reached OD660 of 1.0, and overnight [on]), respectively. ChvR levels were determined by Northern blot analysis; 5S rRNA served as loading control. (C) cDNA reads of +/- MMC libraries mapping to the chvR/recF locus of C. crescentus NA1000. Libraries established from terminator exonuclease (TEX)-treated RNA samples are represented in blue, libraries established from untreated RNA in black, respectively. All libraries were adjusted to the same scale. Annotation and genome position are indicated in the centre. Transcriptional start sites are marked by arrows.