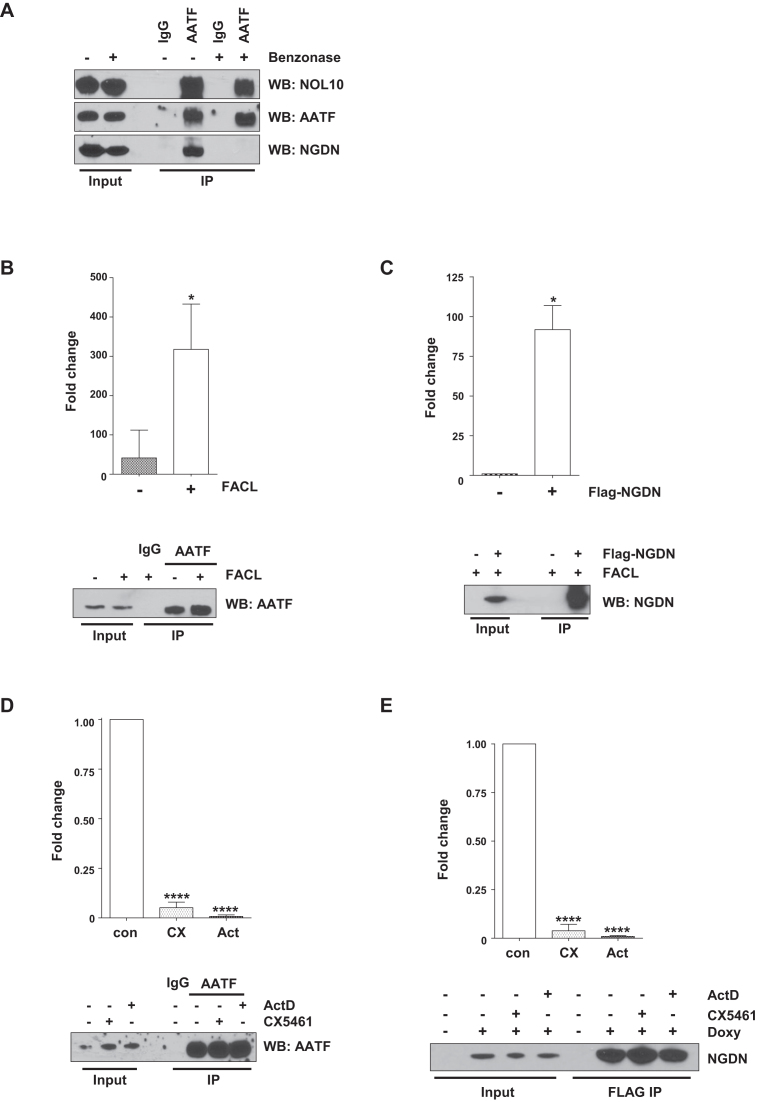
Figure 5.
ANN complex RBPs AATF and NGDN bind to pre-rRNA. (A) AATF was isolated from U-2 OS nuclear extracts by immunoprecipitation using a specific antibody. IgG was used as a negative control. Immunoprecipitations were performed from extracts prepared either with or without Benzonase treatment. The immunoprecipitated proteins were separated by SDS PAGE and AATF, NGDN and NOL10 were detected by western analysis. The data show that AATF forms a complex with NOL10 and NGDN, but that the presence of NGDN in the complex depends on nucleic acid. (B) AATF was immunoprecipitated from nuclear extracts prepared from untreated and formaldehyde cross-linked U-2 OS cells. In addition, control immunoprecipitations using IgG were performed. Samples were subjected to western analysis to confirm the specific isolation of AATF. In parallel, RNA was isolated from immunoprecipitates and RT-qPCR was carried out using primers to detect rRNA precursors. The y-axis on the graph represents the average fold enrichment of rRNA precursors containing the 5′ETS in the different conditions relative to the IgG control. Values are the mean ± SD of three independent experiments. * (P < 0.05) denotes statistically significant differences in RNA levels. (C) Cross-linking RNA immunoprecipitation of NGDN using the NGDN FLAG-tagged cell line. Immunoprecipitation was performed in control (–) and doxycycline-treated (+) cells using the FLAG antibody. Western analysis and qRT-PCR analysis was carried out as described in (B). Y-axis values are the mean ± SD of three independent experiments. * (P < 0.05) denotes statistically significant differences in RNA levels. (D) U-2 OS cells were treated with CX5461 (CX) or ActD (Act), followed by in vivo formaldehyde cross-linking. AATF was immunoprecipitated and protein samples were subjected to western analysis. The RNA isolated from the AATF immunoprecipitations was subjected to RT-qPCR with specific primers as described above. The data represent the mean ± SD of three independent experiments, **** (P < 0.0005). (E) Cross-linking RIP was performed on cells treated as described above, using FLAG antibodies to isolate FLAG-tagged NGDN. Protein samples and RNA samples were processed as described in (D). The data represent the mean ± SD of three independent experiments, **** (P < 0.0005).