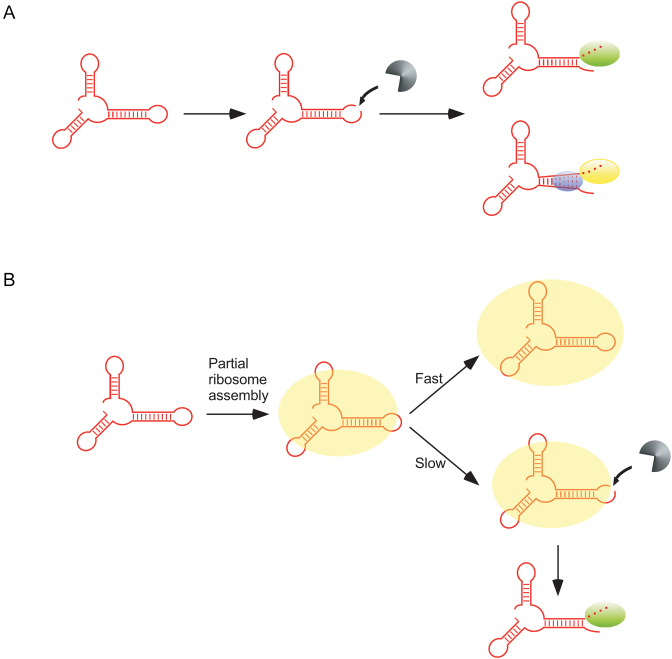
Figure 4.
Two models for the accumulation of rRNA fragments in ΔdeaDΔrnr and ΔsrmBΔrnr strains. (A) A fraction of newly synthesized rRNAs (shown in red) is proposed to be cleaved by an endonuclease, depicted in gray. The resulting products are digested either by RNase R (green oval) or PNPase (yellow oval). PNPase-mediated digestion of structured rRNAs is facilitated via unwinding of base-pairing by SrmB or DeaD (blue ovals). (B) Newly synthesized rRNA is incorporated into a nascent ribosomal particle (show as a yellow oval). In the absence of delays, ribosomal subunit assembly rapidly proceeds to completion, resulting in the protection of the rRNAs from RNase action (top). When ribosome assembly is delayed, endonucleolytic cleavage occurs within exposed rRNA regions (bottom). The resulting fragments are primarily digested by RNase R (green oval) and therefore accumulate in its absence.