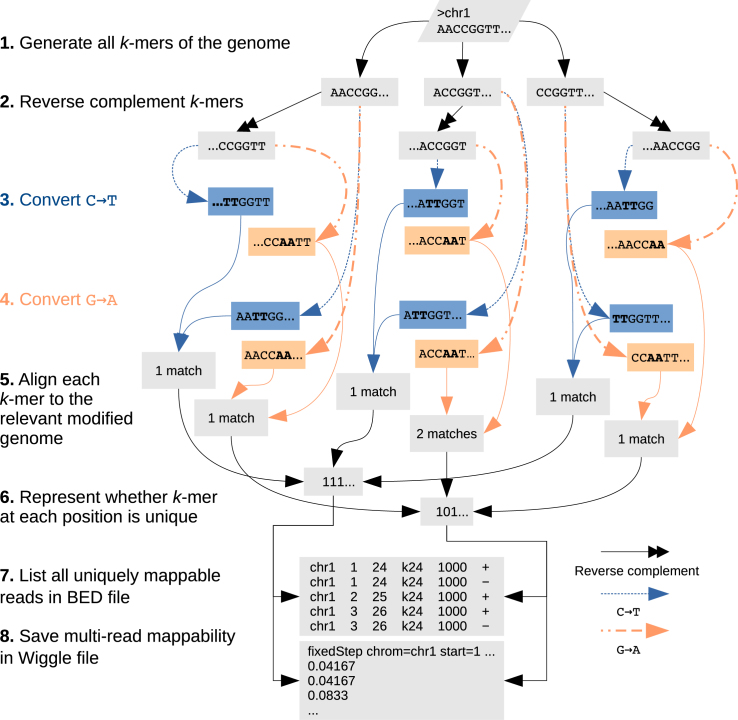
Figure 2.
Mappability of the methylome by Bismap. Bismap identifies uniquely mappable k-mers of a bisulfite-converted genome. It simulates the same changes that may occur in bisulfite treatment on the + strand (C→T) and − strand (G→A). To account for sequence of the − strand, we generate an extra set of reverse-complemented chromosomes and then simulate bisulfite conversion on these chromosomes. We do not simulate reverse complementation after bisulfite conversion, because the experimental protocol does not involve post-conversion DNA amplification. We then align k-mers by disabling complement search and combine the resulting data to quantify the mappability of a bisulfite-converted genome.