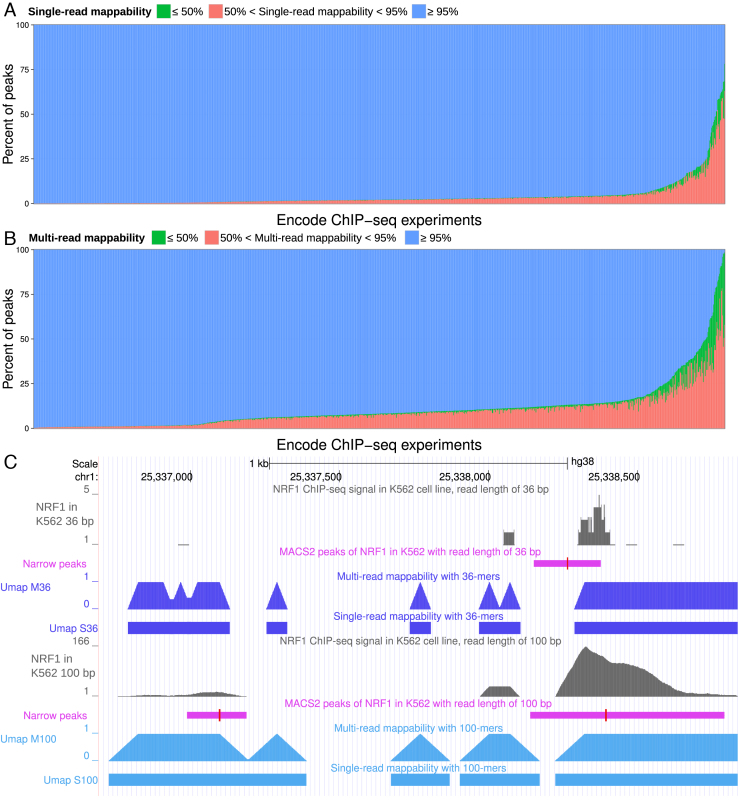
Figure 3.
Mappability of ChIP-seq peaks in 1193 ENCODE datasets. (A) Single-read mappability and (B) multi-read mappability for narrow peaks identified in ENCODE ChIP-seq datasets. (C) An NRF1 narrow peak identified by MACS (purple) that is not uniquely mappable in the experiment with read length of 36 bp. The red bar in peaks indicates the summit. Signal tracks (gray) show two different replicates of this ChIP-seq experiment in K562 chronic myeloid leukemia cells (ENCODE accessions ENCSR000EHH and ENCSR494TDU, with read lengths of 36 and 100 bp, respectively). Umap tracks show single-read and multi-read mappability for two different read lengths of 36 and 100 bp.