Abstract
In Arabidopsis, the circadian clock central oscillator genes are important cellular components to generate and maintain circadian rhythms. There is a negative feedback loop between the morning expressed CCA1 (CIRCADIAN CLOCK ASSOCIATED 1)/LHY (LATE ELONGATED HYPOCOTYL) and evening expressed TOC1 (TIMING OF CAB EXPRESSION 1). CCA1 and LHY negatively regulate the expression of TOC1, while TOC1 also binds to the promoters of CCA1 and LHY to repress their expression. Recent studies indicate that histone modifications play an important role in the regulation of the central oscillators. However, the regulatory relationship between histone modifications and the circadian clock genes remains largely unclear. In this study, we found that the Lysine-Specific Demethylase 1 (LSD1)-like histone demethylases, LDL1 and LDL2, can interact with CCA1/LHY to repress the expression of TOC1. ChIP-Seq analysis indicated that LDL1 targets a subset of genes involved in the circadian rhythm regulated by CCA1. Furthermore, LDL1 and LDL2 interact with the histone deacetylase HDA6 and co-regulate TOC1 by histone demetylation and deacetylaion. These results provide new insight into the molecular mechanism of how the circadian clock central oscillator genes are regulated through histone modifications.
INTRODUCTION
Histone modifications including methylation, acetylation, phosphorylation, ubiquitination and sumoylation play important roles in the regulation of gene expression. All histone modifications are removable, which may therefore provide a flexible way for gene regulation. Methylation on lysine and arginine residues of histone N-terminal tails can be associated with either transcriptional repression or activation. For example, tri-methylation of histone H3 at lysine 4 (H3K4me3) is an active mark for transcription, whereas dimethylation of histone H3 at lysine 9 (H3K9me2) is a signal for transcriptional silencing (1). Histone methylation levels are determined by histone methyltransferases and demethylases, whereas histone acetylation levels are regulated by the action of histone acetyltransferases (HATs) and histone deacetylases (HDACs or HDAs).
Human Lysine-Specific Demethylase 1 (LSD1) is the first histone demethylase identified to demethylate H3K4me through an FAD-dependent oxidation reaction (2). In Arabidopsis, four LSD1 homologs have been identified, namely LSD1-LIKE 1 (LDL1), LDL2, LDL3 and FLOWERING LOCUS D (FLD) (3). In ldl1, ldl1/ldl2 and fld mutant plants, H3K4 methylation on target genes is enriched, suggesting the H3K4 demethylase activity of LDL1, LDL2 and FLD (3,4). In addition, both fld and ldl1/ldl2 double mutant plants show late flowering phenotypes (3,5). FLD represses the expression of FLC by reducing the H3K4me level of FLC chromatin (5,6). Furthermore, LDL1 and LDL2 act redundantly to repress the expression of FLC by H3K4 demethylation. On the other hand, ldl1/ldl2 double mutants also show reduced DNA methylation at the FWA locus, which represses the floral transition (3).
In yeast and animal systems, HDACs and LSD1 are the core components of several multi-protein complexes, such as Mi2/NuRD and CoREST (7–9). HDACs and LSD1 function cooperatively to regulate gene expression in human breast cancer cells (10). Although the HDAC complexes can dynamically interact with different transcription factors depending on different environmental conditions, the interactions between the core protein components in HDAC complexes are relatively stable (11,12). Arabidopsis HDA6 is a RPD3-like class I HDAC involved in transcription repression and regulation of ribosomal RNA (13–17). Furthermore, HDA6 is also involved in flowering, leaf development, senescence and abiotic stress response (6,18–20). Recent studies demonstrated that Arabidopsis HDA6 interacts directly with FLD and affects flowering time by regulating histone H3 acetylation and H3K4 tri-methylation on FLC, MAF4, and MAF5 loci (6).
In Arabidopsis, the circadian clock central oscillators rely on multiple interconnected loops that generate robust rhythms. There is a negative feedback loop between the morning expressed CCA1 (CIRCADIAN CLOCK ASSOCIATED 1) and LHY (LATE ELONGATED HYPOCOTYL) as well as the evening expressed TOC1/PRR1 (TIMING OF CAB EXPRESSION 1/PSEUDO RESPONSE REGULATOR 1). CCA1 and LHY are highly accumulated at dawn, but their expression levels are very low in the evening (21–23). CCA1 and LHY proteins inhibit TOC1 transcription by binding to the evening element (EE) on the TOC1 promoter (21–23). In the evening, TOC1 is highly expressed (23). Recent studies indicate that TOC1 also functions as a repressor and binds to the promoters of CCA1 and LHY to repress their expression in the evening (24,25).
Treating plants with the HDAC inhibitor Trichostatin A (TSA) leads to higher amplitude and delayed phase of TOC1 expression, but the effect of TSA is reduced in toc1 mutant plants (26). In addition, the expression level of CCA1, LHY and TOC1 is specifically associated with changes in the level of H3K4me and H3 acetylation in Arabidopsis (27,28), suggesting that histone modifications are involved in the regulation of circadian central oscillators. Furthermore, the Arabidopsis histone acetyltransferase TAF1 and H3K4 methyltransferase SET DOMAIN GROUP 2/ARABIDOPSIS TRITHORAX RELATED 3 (SDG2/ATXR3) may also contribute directly or indirectly to circadian gene activation (28). In addition, HDA6 functions with PRR9 by interacting with TOPLESS/TOPLESS-RELATED (TPL/TPR) and represses CCA1 transcription (29). These studies indicate that histone modifications play an important role in the regulation of the central oscillators. However, the regulatory relationship between histone modifications and the circadian clock genes remains elusive.
In this study, we found that LDL1/2 interact with the circadian clock central oscillators CCA1/LHY. Furthermore, LDL1/2 interacts with HDA6 and co-regulate TOC1 expression by histone demetylation and deacetylaion. LDL1 targets a subset of genes involved in the circadian rhythm regulated by CCA1. These results provide new insight into the molecular mechanism of how the circadian clock central oscillator TOC1 is repressed through histone modifications.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis (Arabidopsis thaliana) was grown in growth chambers under 12/12 h light/dark conditions at 22°C. In this study, the wild-type Arabidopsis Columbia (Col-0) ecotype was used. The mutants used in this research were previously described, including ldl1/ldl2 (3), hda6 (axe1-5) (6) and cca1/lhy (30). hda6/ldl1/2 triple mutant plants were generated by crossing ldl1/ldl2 and hda6 mutant plants.
Plasmid construction and plant transformation
The full-length coding sequence (CDS) fragments of LDL1, LDL2, CCA1 and LHY were PCR-amplified and cloned into the pCR8/GW/TOPO vector (Invitrogen). HDA6-pCR8/GW/TOPO was described in previous research (6). LDL1 and HDA6 were recombined into the pEarlyGate103-GFP or PK7WGF2 (31) binary vector (Invitrogen), and LDL1, LDL2, CCA1 and LHY were sub-cloned into 3xFLAG Gateway multiple recombination vectors (http://www.psb.ugent/gateway/). Transgenic plants were generated using the floral dip method (32). To construct LDL1-promoter::LDL1:GFP or HDA6-promoter::HDA6:GFP, LDL1 or HDA6 genomic fragment containing ∼2 kb promoter was ligated into the modified pCAMBIA1300 vector containing GFP. pEarlyGate103-LDL1:GFP, PK7WGF2-HDA6:GFP, LDL1pro::LDL1:GFP and HDA6pro::HDA6:GFP were transformed into ldl1 and hda6 by the floral dip method (32).
Bimolecular fluorescence complementation assays
To generate the constructs for BiFC assays, full-length cDNA fragments of LDL1, LDL2, CCA1, LHY, HDA9, HDA18, GLABRA1 (GL1/MYB0) and MYB23 were PCR-amplified and cloned into the pCR8/GW/TOPO vector (Invitrogen), and then recombined into the YN vector pEarleyGate201-YN and the YC vector pEarleyGate202-YC (33). HDA6-YN, HDA6-YC and FLD-YN were described in previous research (6). Constructed vectors were transiently transformed into Arabidopsis protoplasts or tobacco (Nicotiana benthamiana) leaves. Transfected protoplasts and leaves were then examined by using a TCS SP5 confocal spectral microscope imaging system (Leica, https://www.leica.com/).
Yeast two-hybrid and co- immunoprecipitation assays
Yeast two-hybrid assays were performed according to the instruction for the Matchmaker GAL4-based two-hybrid system 3 (Clontech). Full length or truncated LDL1, LDL2, HDA6, CCA1 and LHY cDNA fragments were sub-cloned into pGADT7 and pGBKT7 vectors. All constructs were transformed into the yeast strain AH109 by the lithium acetate method, and yeast cells were grown on a minimal medium/-Leu-Trp according to the manufacturer's instructions (Clontech). Transformed colonies were plated onto a minimal medium/-Leu-Trp-His (3DO) with 0.25 mM 3-amino- 1,2,4-triazole(3AT) or media containing X-α-gal for the α-galactosidase activity assay.
Co-immunoprecipitation assays were performed as previously described (6). Anti-GFP (Santa Cruz Biotechnologies, catalog no. SC-9996; 1:3000 dilution), anti-FLAG (SIGMA catalog no. M2; 1:3000 dilution) and anti-CCA1 (Agrisera, catalog no. AS13 2659; 1:500 dilution) antibodies were used as primary antibodies for Western blot, the resulting signals were detected by using a Pierce ECL Western blotting kit (Pierce, https://www.lifetechnologies.com/).
Quantitative reverse transcriptase PCR analysis
Total RNA was isolated using TRIZOL reagent (Invitrogen, 15596026) according to the manufacturer's instructions. Two micrograms of DNAse (Promega, RQ1 #M6101) treated total RNA were used to synthesize cDNA (Promega, #1012891). RT-qPCR (Real-Time quantitative PCR) was performed using iQ SYBR Green Supermix solution (Bio-Rad, #170-8880). The CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) was used with the following cycling conditions: 95°C for 10 min, followed by 45 cycles of 95°C for 15 s, 60°C for 30 s, and then fluorescent detection. This was immediately followed by a melting curve (65–95°C, incrementing 0.5°C for 5 s, and plate reading). The melting curve analysis confirmed the absence of non-specific products. Each sample was quantified at least in triplicate, and normalized by calculating delta Cq (quantification cycle) to the expression of the internal control Ubiquitin10 (UBQ10). The Cq and relative expression level are calculated by the Biorad CFX Manager 3.1 based on the MIQE guidelines (34). Standard deviations represent at least three technical and two biological replicates. The variance in average data is represented by SEM (standard error of the mean). The SD (standard deviation), SEM determination and P-value were calculated using Student's paired t-test. The gene specific primers used for qRT-PCR are listed in Supplementary Table S1.
Protoplast transient assays
The native promoter driven TOC1pro::LUC plasmid constructs were previously described (30). Effector constructs including 35Spro::TOC1, 35Spro::CCA1, 35Spro::LDL1, 35Spro::HDA6 or 35Spro::GFP were co-transformed into protoplasts with TOC1pro::LUC for transcriptional activity assays, and the plant samples were collected after 12 h at ZT12. The reporter luciferase activities were standardized by activities of co-expressed Renilla luciferase, and relative reporter activities were calculated. Experiments were repeated at least three times for each reporter-effector combination. Signals of Firefly and Renilla luciferase were assayed with the dual luciferase assay reagents (Promega).
Chromatin immunoprecipitation assays
ChIP assays were performed as previously described (6). Chromatin extracts were prepared from seedlings treated with 1% formaldehyde. The chromatin was sheared to the mean length of 500 bp by sonication, proteins and DNA fragments were then immunoprecipitated using antibodies against acetylated histone H3K9K14 (Millipore, catalog no. 06-599), di-methylated histone H3K4 (Diagenode, catalog no. C15410035) or GFP (Abcam, catalog no. ab290). The DNA cross-linked to immunoprecipitated proteins were reversed, and then analyzed by real-time PCR using specific primers (Supplementary Table S1). Percent input was calculated as follows: 2(Cq(IN)-Cq(IP)) × 100. Cq is the quantification cycle as calculated by the Biorad CFX Manager 3.1 based on the MIQE guidelines (34). Standard deviations represent at least three technical and two biological replicates. The variance in average data is represented by SEM (standard error of the mean). The SD (standard deviation), SEM determination and P-value were calculated using Student's paired t-test. The gene specific primers used for real-time PCR are listed in Supplementary Table S1.
ChIP-seq and data analyses
ChIP-seq assays were performed based on previous research (35,36). 10 ng of DNA from at least five ChIPs was pooled to ensure that there are enough starting DNA for library construction. Two biological replicates were prepared and sequenced for each ChIP-seq experiment. The ChIP DNA was first tested by qRT-PCR and then used to prepare ChIP-seq libraries. End repair, adaptor ligation, and amplification were carried out using the Illumina Genomic DNA Sample Prep kit according to the manufacturer's protocol. An Illumina HiSeq 2500 instrument was used for high-throughput sequencing of the ChIP-seq libraries. The raw sequence data were processed using the Illumina sequence data analysis pipeline GAPipeline 1.3.2. Bowtie (37) was then employed to map the reads to the Arabidopsis genome (TAIR10) (38). Only perfectly and uniquely mapped reads were retained for further analysis. To determine the correlation between biological repeats, Pearson correlation was computed using R statistical software on normalized signal intensity for ChIP binding peaks. The alignments were first converted to Wiggle (WIG) files using MACS (39). The data were then imported into the Integrated Genome Viewer (IGV) (40) for visualization. The program SICER (41) was used to identify ChIP-enriched domains (peaks). A peak summit that was positioned within 3 kb upstream or 3 kb downstream of a TSS was assigned to the corresponding gene. If multiple genes could be assigned to a peak, the one with the closest TSS was selected. If no TSS was found in this window, the peak was left unassigned. The overlap venn diagrams were completed by Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html). To identify DNA motifs enriched at LDL1-associated sites, 400-bp sequences encompassing each peak summit (200 bp upstream and 200 bp downstream) were extracted and searched for enriched DNA motifs using MEME-ChIP (42). Searches were performed using default parameters. The binding correlation analysis was completed by Expasy ChIP-Seq tools (http://ccg.vital-it.ch/chipseq/chip_cor.php). The LDL1 ChIP-seq data has been deposited to NCBI-Gene Expression Omnibus (GEO) database (GSE118025). ChIP-Seq files from other research groups, GSE67903 (43), GSE70533 (44) and GSE52175 (un-published ChIP-seq data from Nigel P. D.) were downloaded from the NCBI-GEO database.
RESULTS
LDL1 and LDL2 directly interact with CCA1 and LHY
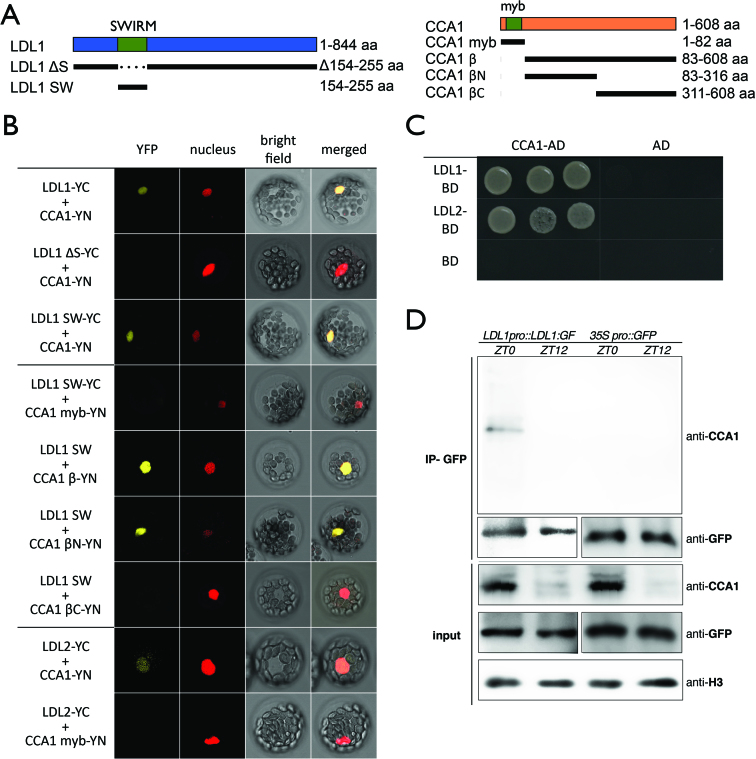
Previous studies indicate that H3K4 methyltransferase activity might be involved in the regulation of circadian central oscillators (28). We analyzed whether the Arabidopsis H3K4 demethylases LDL1 and LDL2 can interact with CCA1 and LHY. Both LDL1 and LDL2 interacted directly with CCA1 and LHY in Bimolecular Fluorescence Complementation (BiFC) assays by using Arabidopsis protoplasts (Figure 1B). Strong YFP signals were observed in the nucleus of the transformed cells. The interactions were further confirmed by yeast two-hybrid (Y2H) assays (Figure 1C, Supplementary Figures S1A and B), Co- immunoprecipitation (Co-IP) assays (Figure 1D, Supplementary Figure S1C) and BiFC assays in Agrobacterium-infiltrated tobacco leaves (Supplementary Figures S1D, S2). The interaction between LDL1 and CCA1 at different timing was analyzed by Co-IP assays using LDL1pro::LDL1:GFP transgenic Arabidopsis plants grown under 12/12 light/dark conditions. Since CCA1 is highly expressed on ZT0, we found that LDL1 interacted with CCA1 on ZT0 but their interaction was abolished on ZT12 (Figure 1D).
Figure 1.
LDL1 and LDL2 interacts with CCA1. (A) Schematic representation of deletions in LDL1 and CCA1 constructs. LDL1ΔS: deletion of 154–255aa; LDL1SW: 154–255aa; CCA1myb: 1–82aa; CCA1β: 83–608aa; CCA1βN: 83–316aa; CCA1βC: 311–608aa. SWIRM: SWIRM domain of LDL1; myb: myb-domain of CCA1. (B) BiFC assays in Arabidopsis protoplasts showing interaction of LDL1/LDL2 with CCA1 in living cells. Different regions of LDL1 and CCA1, and full-length of LDL2 were fused with the N terminus (YN) or C terminus (YC) of YFP and co-delivered into Arabidopsis protoplasts. The nucleus was indicated by mCherry carrying a nuclear localization signal. (C) Yeast two hybrid analysis of the interaction of LDL1/LDL2 with CCA1. LDL1-BD/LDL2-BD with CCA1-AD was co-transformed into the yeast strain AH109. The transformants were plated on the SD/-Leu-Trp-His medium. (D) Co-IP of LDL1:GFP with CCA1 in LDL1pro::LDL1:GFP transformed Arabidopsis. Western blot (WB) was performed with the indicated antibodies.
CCA1β is a splice variant of CCA1, which lacks the MYB-DNA binding domain (45). The N- terminal part of CCA1β contains an important region ( ∼136–316 aa) for its interaction with LHY and for CCA1 dimerization (45,46). To further investigate the interaction domains, various deletion constructs of LDL1 and CCA1 were generated. The interaction was significantly decreased between CCA1 and LDL1 lacking the N-terminal SWIRM domain (47), but the LDL1 SWIRM domain (LDL1SW) can still strongly interact with CCA1 (Figure 1B, Supplementary Figure S3). These results indicated that the SWIRM domain of LDL1 is mainly responsible for the interaction between CCA1 and LDL1. Although full-length LDL1 can interact with both the N-terminal part (CCA1βN, 84–316 aa) and C-terminal part of CCA1β (CCA1βC, 311–608aa), LDL1SW can only interact with CCA1βN (Figure 1B, Supplementary Figures S3 and S4).
LDL1/2 and HDA6 acts synergistically to repress the TOC1 expression by histone deacetylation and H3K4 demethylation
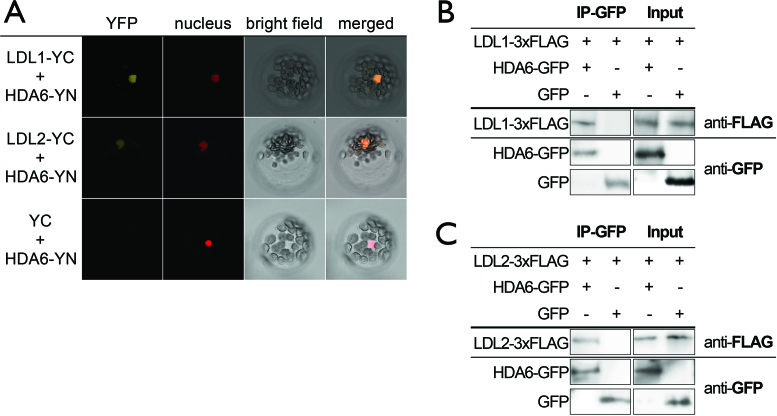
Previous research indicates that Arabidopsis HDA6 directly interacts with the H3K4 demethylase FLD and they synergistically co-regulate gene expression by histone deacetylation and H3K4 demethylation (6). We found that HDA6 can also interact with LDL1 and LDL2 in the nucleus in BiFC assays (Figure 2A, Supplementary Figure S5A). In contrast, LDL1 can not interact with HDA9 and HDA18 in BiFC assays (Supplementary Figure S5A). The interactions of LDL1 and LDL2 with HDA6 were further confirmed by Co-IP assays (Figure 2B and C). These results indicate that LDL1/2 and HDA6 may function in the same protein complex to co-regulate gene expression. This is consistent with previous studies in yeast, animal and plant systems, indicating that HDACs and LSD1-like demethylases function cooperatively to regulate gene expression (6–10). Moreover, HDA6 can also interact with CCA1 and LHY in BiFC assays (Supplementary Figure S5B). In comparison, HDA6 and LDL1 cannot interact with two other Myb transcription factors, GLABRA1 (GL1/MYB0) and MYB23. In addition, FLD cannot interact with CCA1 and LHY in BiFC assays (Supplementary Figure S5B). These results indicate that HDA6 and LDL1 interact with CCA1 and LHY specifically.
Figure 2.
LDL1 and LDL2 interact directly with HDA6. (A) BiFC assays in Arabidopsis protoplasts showing interaction of LDL1/LDL2 with HDA6 in living cells. LDL1/LDL2 and HDA6 fused with the N terminus (YN) or C terminus (YC) of YFP were co-delivered into Arabidopsis protoplasts. The nucleus was indicated by mCherry carrying a nuclear localization signal. (B, C) Co-IP assays of HDA6:GFP with LDL1-3xFLAG (B) or LDL2-3xFLAG (C). Western blot (WB) was performed with the indicated antibodies.
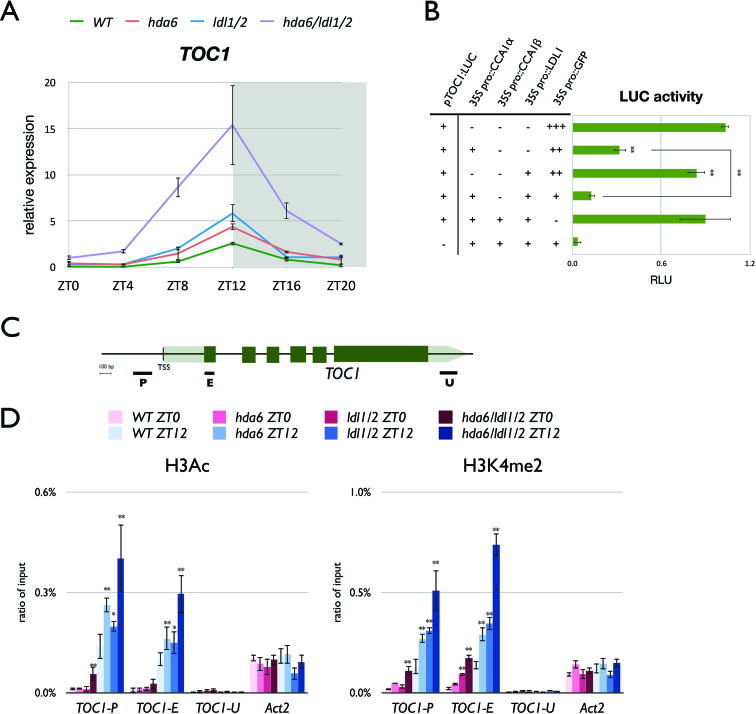
The expression of TOC1 is repressed by CCA1/LHY (22). The interaction of CCA1/LHY with LDL1/2 indicates that they may function together to repress the expression of TOC1. We generated the hda6/ldl1/ldl2 (hda6/ldl1/2) triple mutant by crossing hda6 and ldl1/ldl2 (ldl1/2). Quantitative RT-PCR (qRT-PCR) was used to analyze the daily expression of TOC1 in Columbia (Col-0) wild type (WT), hda6, ldl1/2 and hda6/ldl1/2. The mutant plants were grown under 12 h-light/12 h-dark conditions for 14 days. TOC1 expression was significantly increased in ldl1/2 and hda6 compared to WT (Figure 3A). Furthermore, the expression level of TOC1 was further increased in hda6/ldl1/2 compared to hda6 and ldl1/2 mutants (Figure 3A). However, the expression phase of TOC1 was not shifted in the mutant plants compared to WT (Figure 3A). These results indicate that LDL1/2 and HDA6 may act synergistically to repress the expression of TOC1, although the daily expression phase of TOC1 under 12 h-light/12 h-dark conditions is not affected.
Figure 3.
LDL1/LDL2 and HDA6 acts synergistically to repress TOC1 expression by histone deacetylation and H3K4 demethylation. (A) Gene expression of TOC1 in WT, hda6, ldl1/2 and hda6/ldl1/2. Gene expression was determined by qRT-PCR and normalized to UBQ10. Plants were grown under 12/12 light/dark for 14 days. White and grey regions represent light and dark periods, respectively. (B) Transient luciferase assays in TOC1pro::TOC1:LUC (pTOC1:LUC) transformed protoplasts. The CaMV 35S promoter driven CCA1 (35S pro::CCA1) or LDL1 (35S pro::LDL1) effector constructs were introduced into mesophyll protoplasts extracted from WT. Samples were collected on ZT12 after 12h of transformation. Relative Light Units (RLU) represents firefly luciferase normalized by co-expressed 35Spro::Renilla luciferase. 35Spro::GFP transformed protoplasts were used as a negative control. (C) Schematic diagram of TOC1. P: promoter region, E: coding region, U: 3′ UTR. (D) ChIP analysis of H3K9K14ac and H3K4me2 levels of TOC1. Plants were grown under 12/12 light/dark for 14 days and plant samples were collected on ZT0 and ZT12. The amounts of DNA after ChIP were quantified by qPCR and values represent the average immunoprecipitation efficiencies (%) against the total input DNA. Data points represent average of three technical replicates. Error bars correspond to standard deviations from three biological replicates. *P < 0.05, **P < 0.005 (Student's t-test).
To further confirm whether LDL1 can repress TOC1 expression, the TOC1 promoter driven TOC1 fused with LUCIFERASE (TOC1pro::TOC1:LUC or pTOC1:LUC) was co-expressed with 35Spro::CCA1, 35Spro::LDL1 or 35Spro::GFP in Arabidopsis protoplasts. The LUC expression was only slightly decreased when co-expressed with LDL1, but significantly decreased when co-expressed with LDL1 and CCA1 (Figure 3B). A previous study indicated that co-expression of the full-length CCA1 (CCA1α) with CCA1β inhibits the regulatory function of CCA1 (45). We found that CCA1β can interact with LDL1 and LDL2 in BiFC assays (Figure 1B, Supplementary Figure S3). Interestingly, when LDL1 was co-expressed with CCA1α and CCA1β, the TOC1::LUC expression level was not significantly repressed (Figure 3B). Taken together, these results indicate that CCA1 functions collaboratively with LDL1 to repress TOC1, and the gene repression function of CCA1 and LDL1 was abolished when co-expressed with CCA1β.
To investigate whether CCA1/LHY affect the levels of H3Ac and H3K4me on TOC1, we performed ChIP analysis with WT and cca1/lhy. The levels of H3K4me and H3Ac on TOC1 were significantly increased in cca1/lhy compared to WT (Supplementary Figure S6). Previous studies also indicated that H3K4me and H3Ac on TOC1 were decreased in CCA1 over-expression plants (26,28). We further analyzed whether LDL1, LDL2 and HDA6 are involved in the regulation of the H3K4me and H3Ac levels of TOC1. WT, ldl1/ldl2, hda6 and hda6/ldl1/2 plants grown under 12h-light/12h-dark conditions for 14-days were collected on ZT0 and ZT12 for ChIP-qPCR assays. The levels of H3K4me and H3Ac on the promoter and exon regions of TOC1 were significantly increased in both ldl1/ldl2 and hda6 compared to WT (Figure 3D), indicating that LDL1, LDL2 and HDA6 are indeed involved in TOC1 regulation by removing H3K4me and H3Ac. Furthermore, the H3K4me and H3Ac levels were higher in the hda6/ldl1/2 triple mutant compared to hda6 and ldl1/2 (Figure 3D). Similar results were also obtained with additional putative target genes of LDL1, LDL2 and HDA6 (Supplementary Figure S7). These data support that LDL1, LDL2 and HDA6 act synergistically to change H3K4me and H3Ac states of their targets.
The LDL1-HDA6 complex targets directly to the TOC1 promoter
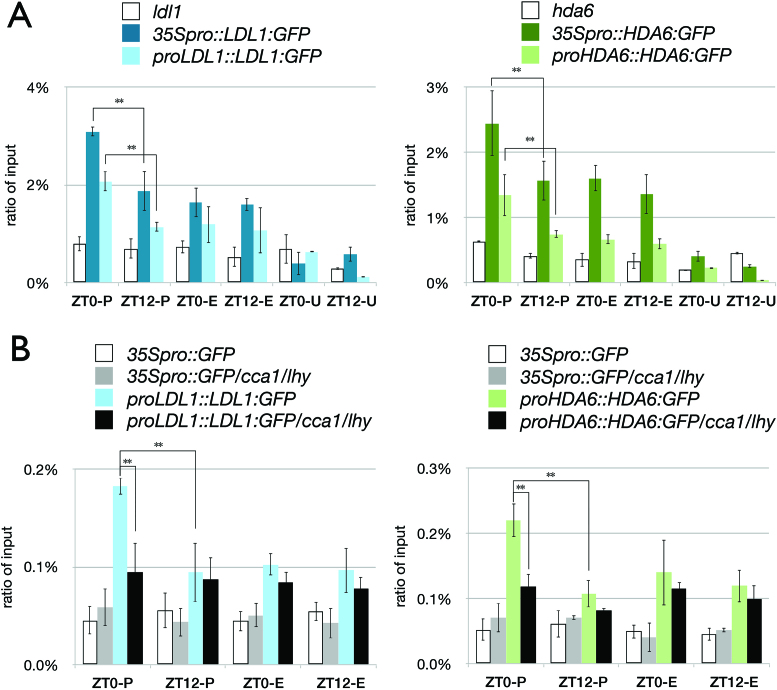
We found that LDL1 interacted with CCA1 on ZT0 but their interaction was abolished on ZT12 (Figure 1D, Supplementary Figure S5C). We further performed ChIP assays to analyze whether LDL1 and HDA6 directly target to TOC1. LDL1:GFP and HDA6:GFP driven by the 35S promoter or their native promoters were transformed into ldl1 and hda6 mutants, respectively. Transgenic seedlings were grown under 12/12 light/dark conditions and harvested on ZT0 and ZT12 after 14 days. ChIP assays were performed with the anti-GFP antibody and the binding of LDL1 and HDA6 to the TOC1 promoter was analyzed by qPCR. As shown in Figure 4, both LDL1 and HDA6 bound to the TOC1 promoter. Furthermore, the binding of LDL1 and HDA6 to the TOC1 promoter was significantly higher on ZT0 than on ZT12 (Figure 4A). The binding of LDL1 and HDA6 to the TOC1 promoter is correlated with the expression of TOC1, since TOC1 is highly expressed in the evening (22).
Figure 4.
Binding of LDL1 and HDA6 to the TOC1 promoter in vivo. (A, B) LDL1 and HDA6 binding to the TOC1 promoter. The schematic diagram of TOC1 is shown in Figure 3. 14 days-old seedlings grown under 12/12 light/dark were harvested on ZT0 or ZT12. ChIP assays were performed with the anti-GFP antibody. The amount of immunoprecipitated DNA was quantified by qRT-PCR. Values represent the average immunoprecipitation efficiencies (%) against the total input DNA. Error bars correspond to standard deviations from three biological replicates. *P < 0.05, **P < 0.005 (Student's t-test).
To analyze whether the binding of LDL1 and HDA6 to the TOC1 promoter is dependent on CCA1 and LHY, LDL1pro::LDL1:GFP and HDA6pro::HDA6:GFP were transformed into the cca1/lhy mutant. The binding of LDL1 and HDA6 on the TOC1 promoter was significantly decreased in cca1/lhy on ZT0 (Figure 4B), supporting that the binding of LDL1 and HDA6 to the TOC1 promoter is dependent on CCA1 and LHY. Although LDL1 and HDA6 also bound to the TOC1 exon regions, this binding was not changed significantly on different time points (Figure 4), indicating that the binding of LDL1 and HDA6 to the TOC1 exon regions may not depend on CCA1/LHY. Previous studies indicated that H3Ac and H3K4me are involved in both transcription initiation and elongation (48,49). The binding of LDL1 and HDA6 to the TOC1 exon region implied that LDL1 and HDA6 may also be involved in both transcription initiation and elongation of TOC1.
LDL1 and CCA1 co-target genes involved in the circadian rhythm
Since most of the identified Arabidopsis circadian clock components display a peak expression at specific times during a day (50,51), we also analyzed the expression patterns of LDL1, LDL2 and HDA6 in Arabidopsis grown under 12/12 light/dark. The expression level of HDA6 was only slightly increased during dark-period, and the daily expression patterns of LDL1 and LDL2 was not changed significantly (Supplementary Figure S8A). In addition, the level of the LDL1 protein did not change significantly on different time points as well (Supplementary Figure S8B). These results indicate that LDL1, LDL2 and HDA6 are continuously expressed.
We further analyzed the expression of the circadian genes in ldl1/ldl2 and hda6/ldl1/2 under ‘free-running’ conditions. Plants grown under 12/12 light/dark for 14 days were transferred to continuous light, and the plant samples were then collected starting from the 2nd day of continuous light. The TOC1 expression phase was shifted on the 2nd day and 3rd day in hda6/ldl1/2. In addition, the expression phase of the circadian marker genes COLD CIRCADIAN RHYTHM AND RNA BINDING 2 (CCR2) and CHLOROPHYLL A/B-BINDING PROTEIN 2 (CAB2) was also shifted in hda6/ldl1/2 on the 2nd or 3rd days of free-running conditions (Supplementary Figure S8D), indicating that LDL1/2 and HDA6 activities contribute to the regulation of circadian rhythm. Previous research also indicated that elevated TOC1 expression causes increased period length (52), which is similar with our results in hda6/ldl1/2. The endogenous circadian clock regulates many physiological output processes including hypocotyl growth. We further analyzed hypocotyl lengths of WT, ldl1/ldl2, hda6 and hda6/ldl1/2. Similar with cca1/lhy, we found that the hypocotyl length of hda6/ldl1/2 was significantly shorter than WT (Supplementary Figure S9). The circadian phenotypes such as period-shift in gene expression and hypocotyl lengths of hda6/ldl1/2 indicate that HDA6 and LDL1/2 may act synergistically to regulate circadian central oscillators.
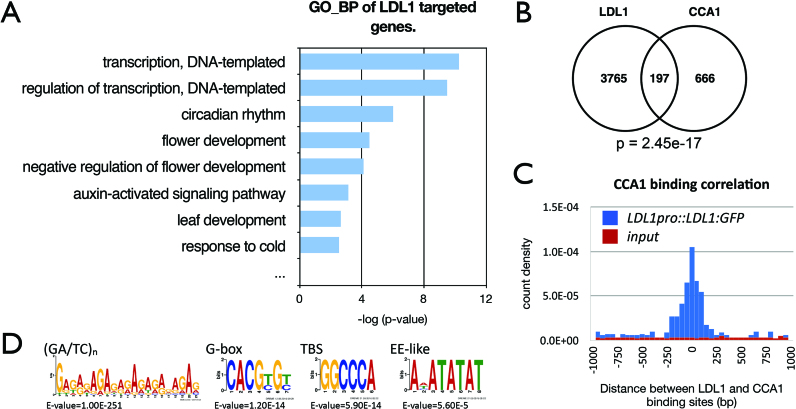
We performed the ChIP sequencing (ChIP-Seq) assays to identify the LDL1-occupied sites in the genome by using the ldl1 plants expressing LDL1pro::LDL1:GFP. Gene Ontology_Biological Process (GO-BP) analysis of LDL1-occupied genes indicated that LDL1 regulates a subset of genes involved in circadian rhythms (Figure 5A, Supplementary Table S2). The newly identified LDL1-occupied genes were compared with the previously published CCA1 ChIP-Seq data, GSE67903 (43). Among 863 genes targeted by CCA1, 197 of them were also occupied by LDL1, indicating a high overlap between LDL1 and CCA1 occupied genes (P = 2.45e–17) (Figure 5B). Gene Ontology (GO) analysis indicates that the LDL1/CCA1 co-targeted genes contain a higher ratio of the circadian rhythm genes compared to the CCA1-targeted or the LDL1-targeted genes alone (Supplementary Figures S10A and B). Moreover, the binding sites of LDL1 and CCA1 were close to each other (Figure 5C), suggesting that LDL1 and CCA1 tend to bind to the same genomic locations. Similar results were also obtained when compared with another published CCA1 ChIP-Seq data, GSE70533 (44) (Supplementary Figures S10C–E). In addition, we also identified a high binding correlation between the LDL1 genomic binding sites and LHY binding sites when compared the LDL1 ChIP-Seq with the LHY ChIP-Seq data, GSE67903 (Supplementary Figure S10F). Previous studies indicate that the cis-elements such as the G-box (CACGTG), TCP binding site (TBS, GGCCCA), Evening Element (EE)-like and (GA/TC)n repeat were enriched in the CCA1-occupied loci (43,44). Similar cis-elements including (GA/TC)n repeat, G-box, TBS and EE-like motifs were also enriched in LDL1-occupied promoter targets (Figure 5D).
Figure 5.
LDL1-occupied sites in the genome identified by ChIP-seq analysis. (A) GO-BP annotation of LDL1 -targeted genes. Annotation terms with P-value < 0.005 were listed. (B) Overlap between CCA1 target genes (43) and LDL1 targeted genes. P = 2.45e–17 (hypergeometric distribution). (C) Distribution of distances between the total binding sites of LDL1 and CCA1. (D) (GA/TC)n, G-box, TBS and EE-like motifs were significantly enriched in the LDL1-binding sites.
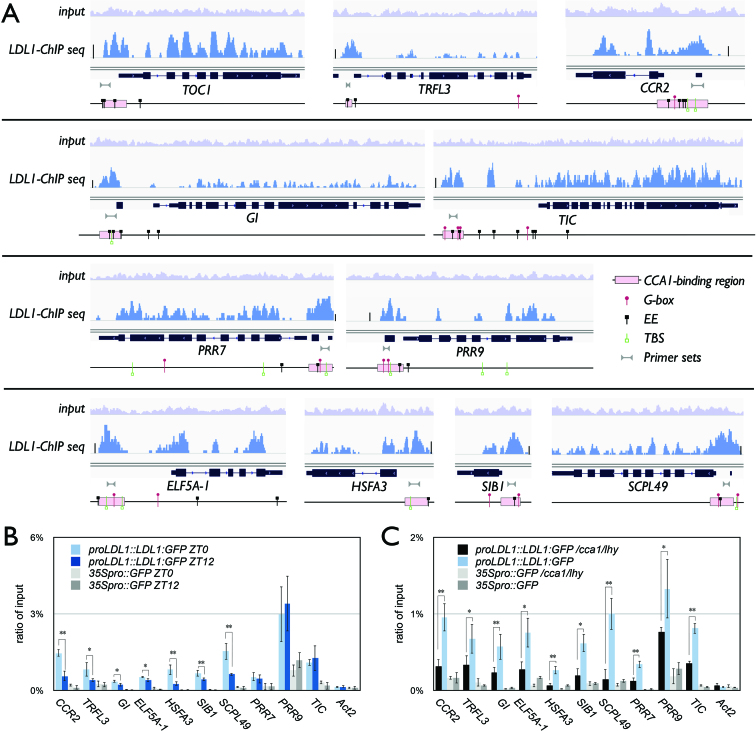
TOC1, PSEUDORESPONSE REGULATOR 7 (PRR7), PRR9, GI, and CCR2 as well as the circadian genes TIME FOR COFFEE (TIC) and TRF-LIKE 3 (TRFL3) were previously identified to be targeted by CCA1 (43,44). These CCA1-occupied genes were also targeted by LDL1. In addition to these genes, we also selected several other genes co-targeted by CCA1/LDL1 for further analysis, including EUKARYOTIC ELONGATION FACTOR 5A-1 (ELF5A-1), HEAT SHOCK TRANSCRIPTION FACTOR A3 (HSFA3), SIGMA FACTOR-BINDING PROTEIN 1 (SIB1) and SERINE CARBOXYPEPTIDASE-LIKE 49 (SCPL49) (43,44) (Figure 6A). Genome browser views of the LDL1 ChIP-Seq data indicate that the LDL1 binding peaks were highly correlated with the CCA1 binding regions and close to the G-box, EE or TBS motifs (Figure 6A). The binding of LDL1 to the promoter regions of these CCA1/LDL1 co-targets was also confirmed by ChIP qRT-PCR (Figure 6B). Similar to TOC1, we found that the binding of LDL1 to the CCR2, TRFL3, GI, ELF5A-1, HSFA3, SIB1 and SCPL49 promoters was significantly higher on ZT0 than ZT12 (Figure 6B). We also analyzed the LDL1 binding to the promoters of the target genes in cca1/lhy mutant plants. The binding of LDL1 on all of the above genes was significantly decreased in cca1/lhy (Figure 6C), suggesting that the binding of LDL1 to promoter regions of the CCA1/LDL1 co-targets are dependent on CCA1 and LHY.
Figure 6.
Binding of LDL1 to the CCA1/LDL1 putative co-targets. (A) Integrated Genome View of LDL1-occupied sites on the CCA1-occupied genes. LDL1 binding peaks of LDL1 and CCA1 putative co-targeted genes are shown. Pink bars indicate the CCA1-binding regions form previous published data (43). Bar summits = 50. (B, C) LDL1 binding to the promoters of CCR2, TRFL3, GI, ELF5A-1, HSFA3, SIB1, SCPL49, PRR7, PRR9 and TIC. 14 days-old seedlings grown under 12/12 light/dark were harvested on ZT0 and ZT12 (B) or ZT0 only (C). ChIP assays were performed with the anti-GFP antibody. The amount of immunoprecipitated DNA was quantified by qRT-PCR. Values represent the average immunoprecipitation efficiencies (%) against the total input DNA. Error bars correspond to standard deviations from three biological replicates. *P < 0.01, **P < 0.001 (Student's t-test).
In addition to TOC1, the expression of other CCA1 targets such as GI and CCR2 was also increased in ldl1/ldl2, hda6 and hda6/ldl1/2 compared to WT (Supplementary Figure S11). The expression of PRR7 and PRR9 was decreased in hda6/ldl1/2 (Supplementary Figure S11). Similarly, the expression of PRR7 and PRR9 was also decreased in cca1/lhy mutants (53). More recent studies suggested that CCA1 functions as a direct transcription repressor of PRR7 and PRR9, but can act indirectly to activate PRR7 and PRR9 in the circadian gene regulatory network (43).
DISCUSSION
There are two types of histone lysine demethylases in eukaryotes, the KDM1/LSD1 and Jumonj C (JmjC) domain-containing demethylases (1,54,55). Four Arabidopsis LSD1 homologs including LDL1, LDL2, LDL3 and FLD have been identified (3). Previous studies indicate that LDL1 and LDL2 act redundantly to repress FLC and FWA expression, and ldl1/ldl2 mutants display a significant late flowering phenotype (3). In this study, we found that LDL1/2 are involved in the regulation of the circadian clock genes. Mis-regulation of the circadian oscillators may result in altered flowering phenotypes, but the precise molecular interaction between the clock and flowering is still poorly understood (51). In addition to LSD1-like type histone demethylases, the Arabidopsis genome encodes 21 JmjC-domain containing proteins, which are capable of removing the methyl-group on different lysine residues (55,56). JMJ30/JMJD5, a JmjC domain-only group protein, was identified as a H3K27me3 demethylase and can regulate the expression of CCA1/LHY (57,58). Arabidopsis RELATIVE OF EARLY FLOWERING 6 (REF6) is a H3K27 demethylase with DNA binding activity (35,59). In addition, the KDM5/JARID1 group JmjC domain-containing proteins were also identified as H3K4 demethylases (55,60,61). The roles of these histone demethylase in regulation of the circadian clock have not been investigated. Furthermore, histone H2B mono-ubiquitination (H2bUb) is also associated with the regulation of CCA1, LHY and TOC1 (62,63), indicating that H2BUb is involved in regulation of circadian central oscillators. The interaction between LDL1 and the H2B deubiquitinase OTU6/OTLD1 has been reported (64). It remains to be determined whether LDL1 and OTU6/OTLD1 act together to regulate the circadian genes.
In yeast and animal systems, HDACs and LSD1 are identified as the core components of several multi-protein complexes such as Mi2/NuRD and CoREST to regulate gene expression cooperatively (7–10). The Arabidopsis LSD1 homolog FLD interacts with HDA6 and regulates flowering (6). In this study, we demonstrate that LDL1/2 can also interact with HDA6 and they act synergistically to regulate gene expression in the circadian clock. Previous studies indicate that HDA6 can be recruited by different transcription factors to regulate gene expression involved in flowering, leaf development, senescence and abiotic stress response (6,12,17–19). In this study, we found that the HDA6-LDL1/2 complex can interact with the Myb-transcription factors CCA1/LHY to regulate TOC1 expression by removing H3Ac and H3K4me on the TOC1 locus. Furthermore, more than 300 genes were co-occupied by LDL1 and CCA1 (43,44). Similar cis-elements such as the G-box, TBS, EE and (GA/TC)n repeat are overrepresented in the binding sites of both CCA1 (43,44) and LDL1, supporting that LDL1 regulates gene expression by interacting with CCA1. CCA1 and LHY are highly expressed in the morning, but their expression levels are very low in the evening (20–22). The interaction between LDL1 and CCA1 was only detectable in the morning in our Co-IP and BiFC analysis. Furthermore, the binding of LDL1 and HDA6 to the TOC1 promoter was also decreased in the evening. CCA1/LHY can therefore recruit the histone modification complex containing LDL1/2 and HDA6 to repress the expression of TOC1 in the morning. However, only about 20% of the putative CCA1-targeted genes were co-targeted by LDL1 in our ChIP-seq experiment, suggesting that maybe not all CCA1 targeted genes are co-regulated by LDL1.
Arabidopsis circadian clock genes generate expression rhythms by multiple interconnected loops and form a complicate feedback regulation network. The central oscillators CCA1, LHY and TOC1 constitute the central loop (20–23). The central loop is meshed with the morning loop and the evening loop, in which the morning loop contains CCA1/LHY, PRR5, PRR7, and PRR9 (65,66), whereas the evening loop is consist of GI, ZEITLUPE (ZTL), PRR3 and TOC1 (67–70). We found that although the binding of LDL1 and HDA6 to the TOC1 promoter region was decreased in the cca1/lhy mutant plants, their binding was not completely abolished, suggesting that other unknown transcription factors may also recruit the LDL1/2-HDA6 complex to TOC1. In addition to CCA1/LHY, the Evening Complex (EC) comprising of EARLY FLOWERING3 (ELF3), EARLY FLOWERING4 (ELF4) and the small putative Myb transcription factor LUX ARRHYTHMO (LUX) is also involved in the regulation of TOC1 (71,72), since TOC1 expression is increased in elf3, elf4 and lux (73–75). It was reported that HDA6 functions with PRR9 by interacting with TOPLESS/TOPLESS-RELATED (TPL/TPR) to repress CCA1 (29). TOC1 (also known as PRR1) is one of the PRR-family proteins (76). The functional correlation of LDL1/2-HDA6 with TOC1, PRRs and EC remains to be further clarified. By using the ChIP-Seq assay, we identified additional circadian genes including the central loop, morning loop and evening loop genes are targeted by LDL1. Since the morning loop and evening loop circadian genes are also involved in the feedback regulation of CCA1 and LHY, it remains to be determined whether this feedback regulation is functional related to LDL1/2 and HDA6.
Recent studies indicated that the circadian clock genes including TOC1 and PRRs are regulated by alternative splicing of RNA transcripts (77–79). While histone acetylation and methylation are important for fine-tuning the accessibility of chromatin, it is believed that pre-mRNA splicing can combine with histone modification events. The association of H3K4me and H3Ac with alternative splicing has been demonstrated in mammalian systems (49,80). In this research, we found that LDL1 and HDA6 can bind to the gene-body of TOC1. The ChIP-seq data also showed that LDL1 can bind to the gene-bodies of many circadian genes. These results implied that the LDL1/2-HDA6 complex may be involved in the other regulation processes such as transcription elongation and alternative splicing regulation of the circadian genes.
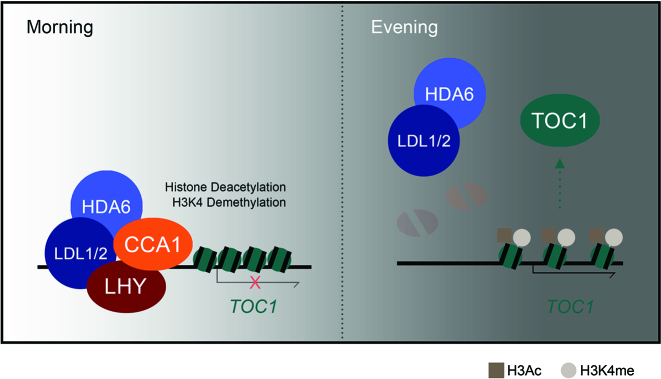
In summary, we demonstrated how LDL1/2 and HDA6 are involved in the regulation of the circadian central oscillators TOC1 by histone deacetylation and H3K4 demethylation (Figure 7). The morning accumulated-CCA1/LHY interact with the histone modification complex containing LDL1/2 and HDA6. CCA1/LHY act as transcription repressors and recruit the LDL1/2-HDA6 histone modification complex to their targets locus such as TOC1. In the evening, CCA1/LHY are low expressed, and TOC1 is highly expressed because the LDL1/2-HDA6 complex is released from the TOC1 promoter.
Figure 7.
A model for LDL1/2 and HDA6 functions in the regulation of circadian clock central oscillator TOC1. The morning accumulated-CCA1/LHY interact with the histone modification complex containing LDL1/2 and HDA6. CCA1/LHY act as transcription repressors and recruit the histone modification complex to their targets loci such as TOC1. In the evening, CCA1/LHY are low expressed, and TOC1 is highly expressed because the LDL1/2-HDA6 complex is released from the TOC1 promoter.
DATA AVAILABILITY
The LDL1 ChIP-seq data was deposited to NCBI-Gene Expression Omnibus (GEO) database (GSE118025).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. S.-H. Wu. and J.-F. Wu for sharing the cca1/lhy double mutant and TOC1pro::LUC construct. We also thank Technology Commons, College of Life Science, National Taiwan University for the convenient use of the Bio-Rad real-time PCR system and the confocal spectral microscope imaging systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of the Republic of China [105-2311-B-002-012-MY3 and 106-2313-B-002-003- to K.W.]; National Taiwan University [106R891501 and 107L893101 to K.W.]; Agriculture and Agri-Food Canada A-base and the National Science and Engineering Research Council of Canada [RGPIN/04625-2017 to Y.C.]. Funding for open access charge: National Taiwan University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lan F., Nottke A.C., Shi Y.. Mechanisms involved in the regulation of histone lysine demethylases. Curr. Opin. Cell Biol. 2008; 20:316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y.. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004; 119:941–953. [DOI] [PubMed] [Google Scholar]
- 3. Jiang D., Yang W., He Y., Amasino R.M.. Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell. 2007; 19:2975–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Greenberg M.V.C., Deleris A., Hale C.J., Liu A., Feng S.H., Jacobsen S.E.. Interplay between active chromatin marks and RNA-Directed DNA methylation in Arabidopsis thaliana. PLos Genet. 2013; 9:e1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He Y., Michaels S.D., Amasino R.M.. Regulation of flowering time by histone acetylation in Arabidopsis. Science. 2003; 302:1751–1754. [DOI] [PubMed] [Google Scholar]
- 6. Yu C.W., Liu X., Luo M., Chen C., Lin X., Tian G., Lu Q., Cui Y., Wu K.. HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 2011; 156:173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khochbin S., Verdel A., Lemercier C., Seigneurin-Berny D.. Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 2001; 11:162–166. [DOI] [PubMed] [Google Scholar]
- 8. Lee M.G., Wynder C., Cooch N., Shiekhattar R.. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005; 437:432–435. [DOI] [PubMed] [Google Scholar]
- 9. Wang Y., Zhang H., Chen Y., Sun Y., Yang F., Yu W., Liang J., Sun L., Yang X., Shi L et al. . LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009; 138:660–672. [DOI] [PubMed] [Google Scholar]
- 10. Huang P.H., Chen C.H., Chou C.C., Sargeant A.M., Kulp S.K., Teng C.M., Byrd J.C., Chen C.S.. Histone deacetylase inhibitors stimulate histone H3 lysine 4 methylation in part via transcriptional repression of histone H3 lysine 4 demethylases. Mol. Pharmacol. 2011; 79:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Joshi P., Greco T.M., Guise A.J., Luo Y., Yu F., Nesvizhskii A.I., Cristea I.M.. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol. 2013; 9:672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu X., Yang S., Zhao M., Luo M., Yu C.W., Chen C.Y., Tai R., Wu K.. Transcriptional repression by histone deacetylases in plants. Mol Plant. 2014; 7:764–772. [DOI] [PubMed] [Google Scholar]
- 13. Murfett J., Wang X.J., Hagen G., Guilfoyle T.J.. Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell. 2001; 13:1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Probst A.V., Fagard M., Proux F., Mourrain P., Boutet S., Earley K., Lawrence R.J., Pikaard C.S., Murfett J., Furner I. et al. . Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell. 2004; 16:1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Earley K., Lawrence R.J., Pontes O., Reuther R., Enciso A.J., Silva M., Neves N., Gross M., Viegas W., Pikaard C.S.. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006; 20:1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu X.C., Yu C.W., Duan J., Luo M., Wang K.C., Tian G., Cui Y.H., Wu K.Q.. HDA6 directly Interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 2012; 158:119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu C.W., Tai R., Wang S.C., Yang P., Luo M., Yang S., Cheng K., Wang W.C., Cheng Y.S., Wu K.. HISTONE DEACETYLASE6 acts in concert with histone methyltransferases SUVH4, SUVH5, and SUVH6 to regulate transposon silencing. Plant Cell. 2017; 29:1970–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu K., Zhang L., Zhou C., Yu C.W., Chaikam V.. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 2008; 59:225–234. [DOI] [PubMed] [Google Scholar]
- 19. Chen L.T., Luo M., Wang Y.Y., Wu K.. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 2010; 61:3345–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luo M., Yu C.W., Chen F.F., Zhao L., Tian G., Liu X., Cui Y., Yang J.Y., Wu K.. Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in arabidopsis. PLos Genet. 2012; 8:e1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carre I.A., Coupland G.. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998; 93:1219–1229. [DOI] [PubMed] [Google Scholar]
- 22. Wang Z.Y., Tobin E.M.. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998; 93:1207–1217. [DOI] [PubMed] [Google Scholar]
- 23. Alabadi D., Oyama T., Yanovsky M.J., Harmon F.G., Mas P., Kay S.A.. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001; 293:880–883. [DOI] [PubMed] [Google Scholar]
- 24. Gendron J.M., Pruneda-Paz J.L., Doherty C.J., Gross A.M., Kang S.E., Kay S.A.. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang W., Perez-Garcia P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P.. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012; 336:75–79. [DOI] [PubMed] [Google Scholar]
- 26. Perales M., Mas P.. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell. 2007; 19:2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malapeira J., Khaitova L.C., Mas P.. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:21540–21545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hemmes H., Henriques R., Jang I.C., Kim S., Chua N.H.. Circadian clock regulates dynamic chromatin modifications associated with Arabidopsis CCA1/LHY and TOC1 transcriptional rhythms. Plant Cell Physiol. 2012; 53:2016–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang L., Kim J., Somers D.E.. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y., Wu J.F., Nakamichi N., Sakakibara H., Nam H.G., Wu S.H.. LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell. 2011; 23:486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karimi M., Inze D., Depicker A.. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002; 7:193–195. [DOI] [PubMed] [Google Scholar]
- 32. Clough S.J., Bent A.F.. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998; 16:735–743. [DOI] [PubMed] [Google Scholar]
- 33. Lu Q., Tang X., Tian G., Wang F., Liu K., Nguyen V., Kohalmi S.E., Keller W.A., Tsang E.W., Harada J.J. et al. . Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 2010; 61:259–270. [DOI] [PubMed] [Google Scholar]
- 34. Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. et al. . The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009; 55:611–622. [DOI] [PubMed] [Google Scholar]
- 35. Li C.L., Gu L.F., Gao L., Chen C., Wei C.Q., Qiu Q., Chien C.W., Wang S.K., Jiang L.H., Ai L.F. et al. . Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 2016; 48:687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li C.L., Chen C., Gao L., Yang S.G., Nguyen V., Shi X.J., Siminovitch K., Kohalmi S.E., Huang S.Z., Wu K.Q. et al. . The Arabidopsis SWI2/SNF2 chromatin remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor Gene SVP. PLos Genet. 2015; 11:e1004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M. et al. . The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012; 40:D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W.. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009; 25:1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Machanick P., Bailey T.L.. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011; 27:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kamioka M., Takao S., Suzuki T., Taki K., Higashiyama T., Kinoshita T., Nakamichi N.. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell. 2016; 28:696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nagel D.H., Doherty C.J., Pruneda-Paz J.L., Schmitz R.J., Ecker J.R., Kay S.A.. Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4802–E4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Seo P.J., Park M.J., Lim M.H., Kim S.G., Lee M., Baldwin I.T., Park C.M.. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell. 2012; 24:2427–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu S.X., Knowles S.M., Andronis C., Ong M.S., Tobin E.M.. CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009; 150:834–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Aravind L., Iyer L.M.. The SWIRM domain: a conserved module found in chromosomal proteins points to novel chromatin-modifying activities. Genome Biol. 2002; 3:RESEARCH0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao Y.M., Lu J., Sun H., Chen X., Huang W.F., Tao D., Huang B.Q.. Histone acetylation regulates both transcription initiation and elongation of hsp22 gene in Drosophila. Biochem. Biophys. Res. Commun. 2005; 326:811–816. [DOI] [PubMed] [Google Scholar]
- 49. He R.S., Kidder B.L.. H3K4 demethylase KDM5B regulates global dynamics of transcription elongation and alternative splicing in embryonic stem cells. Nucleic Acids Res. 2017; 45:6427–6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hazen S.P., Naef F., Quisel T., Gendron J.M., Chen H.M., Ecker J.R., Borevitz J.O., Kay S.A.. Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol. 2009; 10:R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nagel D.H., Kay S.A.. Complexity in the wiring and regulation of plant circadian networks (vol 22, pg R648, 2012). Curr. Biol. 2013; 23:95–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mas P., Alabadi D., Yanovsky M.J., Oyama T., Kay S.A.. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell. 2003; 15:223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Farre E.M., Harmer S.L., Harmon F.G., Yanovsky M.J., Kay S.A.. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 2005; 15:47–54. [DOI] [PubMed] [Google Scholar]
- 54. Klose R.J., Kallin E.M., Zhang Y.. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006; 7:715–727. [DOI] [PubMed] [Google Scholar]
- 55. Luo M., Hung F.Y., Yang S.G., Liu X.C., Wu K.Q.. Histone Lysine Demethylases and Their Functions in Plants. Plant Mol. Biol. Rep. 2014; 32:558–565. [Google Scholar]
- 56. Lu F.L., Li G.L., Cui X., Liu C.Y., Wang X.J., Cao X.F.. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J. Integr. Plant Biol. 2008; 50:886–896. [DOI] [PubMed] [Google Scholar]
- 57. Jones M.A., Covington M.F., DiTacchio L., Vollmers C., Panda S., Harmer S.L.. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21623–21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gan E.S., Xu Y.F., Wong J.Y., Goh J.G., Sun B., Wee W.Y., Huang J.B., Ito T.. Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat. Commun. 2014; 5:5098. [DOI] [PubMed] [Google Scholar]
- 59. Lu F.L., Cui X., Zhang S.B., Jenuwein T., Cao X.F.. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 2011; 43:715–719. [DOI] [PubMed] [Google Scholar]
- 60. Jeong J.H., Song H.R., Ko J.H., Jeong Y.M., Kwon Y.E., Seol J.H., Amasino R.M., Noh B., Noh Y.S.. Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS One. 2009; 4:e8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang H., Mo H., Fan D., Cao Y., Cui S., Ma L.. Overexpression of a histone H3K4 demethylase, JMJ15, accelerates flowering time in Arabidopsis. Plant Cell Rep. 2012; 31:1297–1308. [DOI] [PubMed] [Google Scholar]
- 62. Himanen K., Woloszynska M., Boccardi T.M., De Groeve S., Nelissen H., Bruno L., Vuylsteke M., Van Lijsebettens M.. Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J. 2012; 72:249–260. [DOI] [PubMed] [Google Scholar]
- 63. Bourbousse C., Ahmed I., Roudier F., Zabulon G., Blondet E., Balzergue S., Colot V., Bowler C., Barneche F.. Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLos Genet. 2012; 8:e1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Krichevsky A., Zaltsman A., Lacroix B., Citovsky V.. Involvement of KDM1C histone demethylase-OTLD1 otubain-like histone deubiquitinase complexes in plant gene repression. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:11157–11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.H., Sakakibara H.. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010; 22:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Salome P.A., Weigel D., McClung C.R.. The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell. 2010; 22:3650–3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim W.Y., Geng R., Somers D.E.. Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:4933–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Para A., Farre E.M., Imaizumi T., Pruneda-Paz J.L., Harmon F.G., Kay S.A.. PRR3 Is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell. 2007; 19:3462–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. McClung C.R., Gutierrez R.A.. Network news: prime time for systems biology of the plant circadian clock. Curr. Opin. Genet. Dev. 2010; 20:588–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mas P., Kim W.Y., Somers D.E., Kay S.A.. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003; 426:567–570. [DOI] [PubMed] [Google Scholar]
- 71. Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farre E.M., Kay S.A.. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011; 475:398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pokhilko A., Fernandez A.P., Edwards K.D., Southern M.M., Halliday K.J., Millar A.J.. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012; 8:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kolmos E., Nowak M., Werner M., Fischer K., Schwarz G., Mathews S., Schoof H., Nagy F., Bujnicki J.M., Davis S.J.. Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J. 2009; 3:350–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dixon L.E., Knox K., Kozma-Bognar L., Southern M.M., Pokhilko A., Millar A.J.. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 2011; 21:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hazen S.P., Schultz T.F., Pruneda-Paz J.L., Borevitz J.O., Ecker J.R., Kay S.A.. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci U.S.A. 2005; 102:10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Matsushika A., Makino S., Kojima M., Mizuno T.. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol. 2000; 41:1002–1012. [DOI] [PubMed] [Google Scholar]
- 77. Jones M.A., Williams B.A., McNicol J., Simpson C.G., Brown J.W., Harmer S.L.. Mutation of Arabidopsis spliceosomal timekeeper locus1 causes circadian clock defects. Plant Cell. 2012; 24:4066–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Perez-Santangelo S., Schlaen R.G., Yanovsky M.J.. Genomic analysis reveals novel connections between alternative splicing and circadian regulatory networks. Brief. Funct. Genomics. 2013; 12:13–24. [DOI] [PubMed] [Google Scholar]
- 79. Perez-Santangelo S., Mancini E., Francey L.J., Schlaen R.G., Chernomoretz A., Hogenesch J.B., Yanovsky M.J.. Role for LSM genes in the regulation of circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15166–15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shindo Y., Nozaki T., Saito R., Tomita M.. Computational analysis of associations between alternative splicing and histone modifications. FEBS Lett. 2013; 587:516–521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The LDL1 ChIP-seq data was deposited to NCBI-Gene Expression Omnibus (GEO) database (GSE118025).