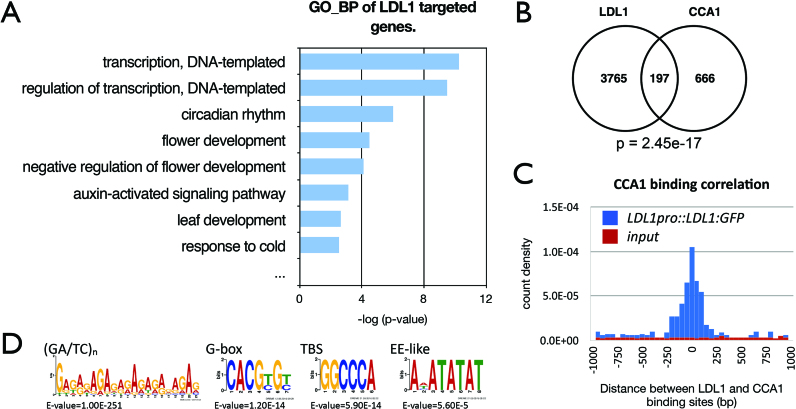
Figure 5.
LDL1-occupied sites in the genome identified by ChIP-seq analysis. (A) GO-BP annotation of LDL1 -targeted genes. Annotation terms with P-value < 0.005 were listed. (B) Overlap between CCA1 target genes (43) and LDL1 targeted genes. P = 2.45e–17 (hypergeometric distribution). (C) Distribution of distances between the total binding sites of LDL1 and CCA1. (D) (GA/TC)n, G-box, TBS and EE-like motifs were significantly enriched in the LDL1-binding sites.