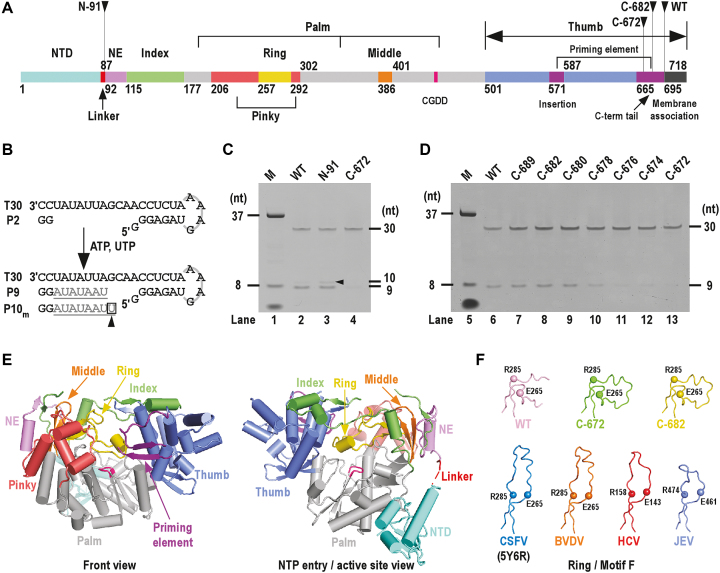
Figure 1.
Functional/structural element assignment, the de novo RdRP activity assessment and the global structure of CSFV NS5B. ( A) A color-coded bar of functional or structural elements of CSFV NS5B. Coloring scheme: NTD in teal, linker in red, RdRP palm in grey, N-terminal extension (NE) in violet, index finger in green, middle finger in orange, ring finger in yellow, pinky finger in light red, thumb in slate, priming element in purple, motif C signature sequence CGDD in magenta and membrane association region in black. The numbers defining the residue ranges of each element are shown. (B) A schematic diagram of the de novo-mode RdRP assays. Construct T30/P2 was used as the RNA substrate. When ATP and UTP were supplied as the only NTP substrates, the template-strand T30 directed a 7-nucleotide (grey) extension of the dinucleotide primer P2 (black) to produce a 9-mer product (P9). The 10-mer product was generated through a G:Umis event. (C and D) Comparison of the de novo RdRP activity for the WT NS5B and its N-/C-terminal truncated forms. The oval-shaped band below the 8-nt marker (M) is the bromphenol blue mixed with the marker sample. Note that the 8-nt marker was chemically synthesized and bearing hydroxyl groups at the 5′-end, and therefore migrated slower than the 9-nt product bearing a 5′-phosphate. (E) Global views of CSFV NS5B crystal structure. Structure of NS5B C-682 construct shown in orientations viewing into the front channel (left) and NTP entry channel (right). The coloring scheme is consistent with that in panel-A. (F) A structural comparison of the Flaviviridae RdRP ring finger (motif F). The ring finger is shown as noodles with the α-carbon atoms of two highly conserved motif F residues in spheres. Top row: CSFV NS5B constructs; bottom row: representative Flaviviridae NS5B constructs. PDB entries: 5YF5 (WT); 5YF6 (C-682); 5YF7 (C-672); 5Y6R (CSFV) (42); 1S4F (BVDV) (39); 1NB4 (HCV) (59); 4K6M (JEV) (41).