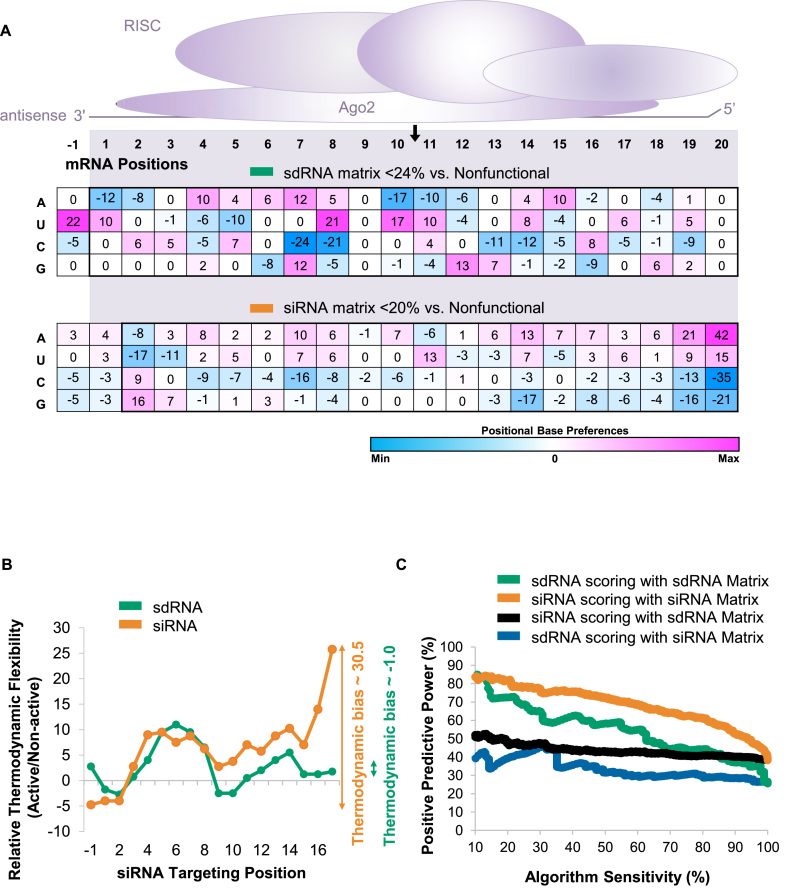
Figure 4.
Algorithms derived from naked siRNA do not have predictive power for modified (sdRNAs) and vice versa. (A) The positional base preference matrix was generated from non-modified (orange) (5) and chemically modified (green) sdRNA. Sequences were aligned based on the 5′ end of the antisense strand. Matrix weight values are color-coded by value as indicated by shaded bar below the matrices. Analyzed mRNA positions corresponding to siRNA-targeting region (shaded) are indicated at the top. Black arrow indicates the location of cleavage site between positions 10 and 11. (B) The ability of algorithms derived from non-modified and modified sdRNA datasets to predict the efficacy of non-modified and modified siRNAs was calculated using PPP vs sensitivity plots. (C) The thermodynamic flexibility of the non-modified and chemically modified siRNAs was estimated by averaging GC content over a sliding window of four bases. Thermodynamic bias is indicated as the difference between the relative thermodynamic flexibility at 5′ and 3′ ends of the siRNA duplex. Chemically modified siRNAs do not display conventional thermodynamic bias.