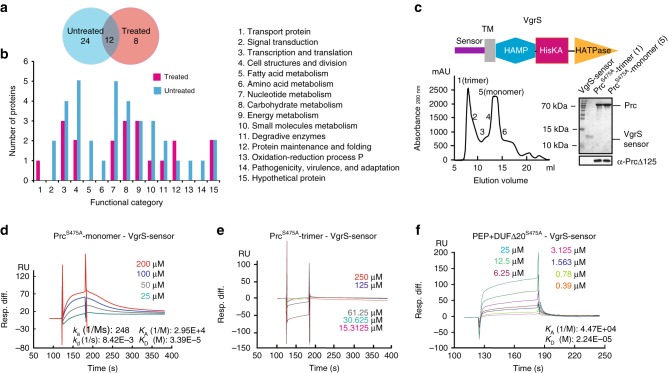
Fig. 3.
Tandem affinity purification (TAP) identify that the VgrS sensor directly binds the Prc monomer. a Venn diagram of the number of proteins identified by TAP together with a nanoLC–MS/MS analysis. Samples subjected or not subjected to osmostress were analysed. b Functional categories of the putative Prc binding proteins. Protein details are listed in Supplementary Table 3. c Prc exists as a monomer and a trimer in vitro. Upper panel: Secondary structure of VgrS. Lower left panel: Recombinant PrcS475A was separated by a molecular sieve, and the molecular weights of the fractions were measured by analytical ultracentrifugation. Lower right panel: Purification of the VgrS sensor and Prc proteins. Western blotting was used to verify the Prc proteins, as shown below. d–f Quantification of the binding affinity between the VgrS sensor and recombinant Prc by surface plasmon resonance. d The VgrS sensor bound the PrcS475A monomer. e The VgrS sensor did not bind the PrcS475A trimer. f The VgrS sensor bound a truncated PrcS475A, which contains peptidase and DUF3340 domains. The VgrS sensor protein was trapped on a sensor CM5 chip, and various concentrations of Prc were injected at a flow-rate of 30 μl/min at 25 °C. Data were analysed using a model for a single set of identical binding sites. The binding kinetics of the Prc–VgrS sensor interaction: ka association rate constant; kd dissociation rate constant; KA equilibrium association rate constant; and KD equilibrium dissociation rate constant