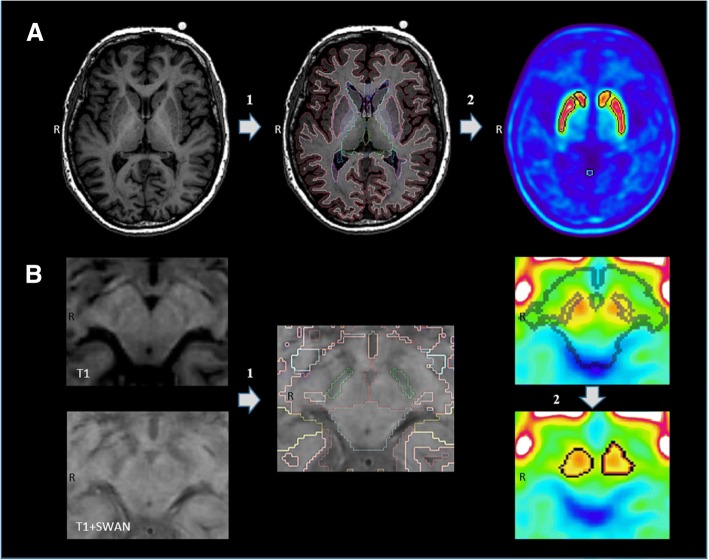
Fig. 3.
Illustration of the procedure for PET image analysis. a Measurement of the FE-PE2I activity in the caudate and the putamen: In the first step (1), T1-weighed MRI data was automatically segmented using Freesurfer, delineating the structural borders of the caudate and putamen in each hemisphere. In the second step (2), delineation of the PET-data was done, restricted by the limits of the segmented VOIs created in the first step. New PET-based VOIs were delineated for the caudate and putamen, defined as the voxels with the highest activity within a volume of 2.0 and 2.5 mL respectively. This was done to adjust for imprecise Freesurfer MRI segmentation, due to periventricular white matter irregularities, and to minimize partial volume effects in the PET image data. b Measurement of the FE-PE2I activity in the SN. In the first step (1), manual segmentation was anatomically guided by the hypo-intense area in the midbrain corresponding to SN on merged SWAN and T1-weighted MRI transversal slices. Since the MRI-guided VOIs did not entirely overlap the area of the observed FE-PE2I binding in the midbrain, in the second step (2), these segmented VOIs were used to guide an automatic thresholding algorithm to determine PET-based midbrain VOIs. FE-PE2I, [18F]FE-PE2I PET; MRI, magnetic resonance imaging; VOI, volume of interest; SN, substantia nigra; SWAN, susceptibility-weighted angiography