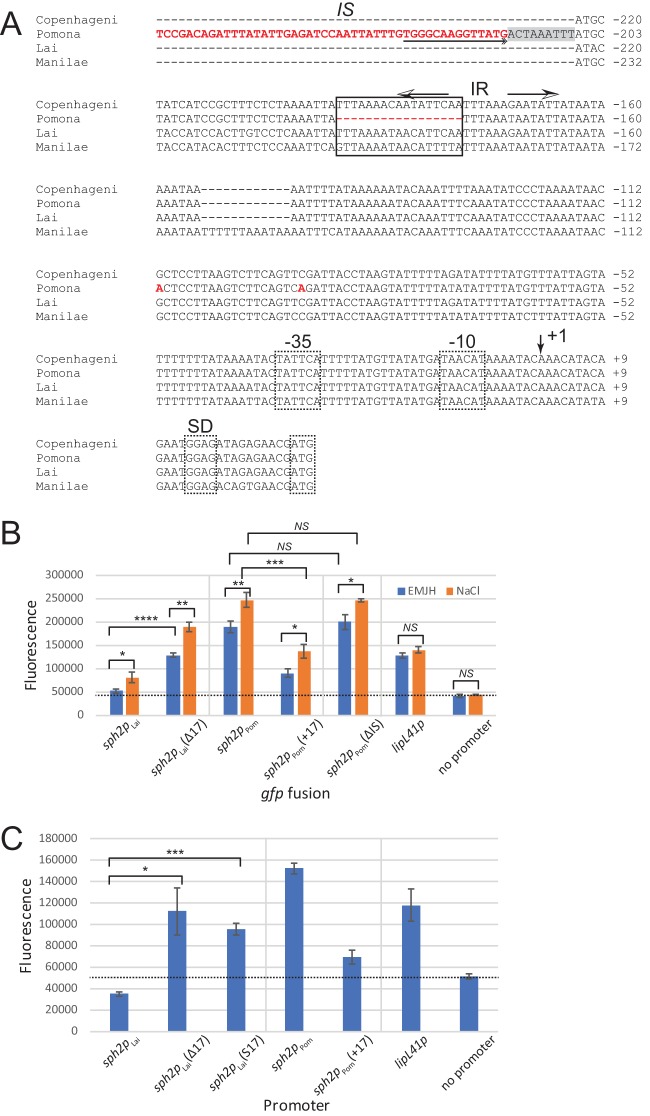
FIG 8.
Effect of mutations introduced upstream of the sph2 promoter. (A) Multisequence alignment. Sequences upstream of sph2 from L. interrogans strains Fiocruz L1-130 (Copenhageni), LC82-25 (Pomona), 56601 (Lai), and L495 (Manilae) were aligned with Kalign. The aligned sequences extend from the 5′ ends of the promoter fragments cloned into pRAT724 to the start codon of sph2. Most of the IS-like sequences and sequences further upstream were omitted from the figure for brevity. The sequence and nucleotides unique to the Pomona LC82-25 are shown in red. An outside end of the IS-like element containing an inverted repeat is underlined with a double arrowhead at the end. The duplicated host sequence adjacent to the insertion site of the IS-like element is shaded gray. The key 17-nucleotide sequence is boxed, and the imperfect inverted repeats (IR) overlapping with the key sequence are marked with arrows. The proposed transcription start site is indicated by an arrow at position +1. The E. coli-like −10 and −35 promoter sequences, Shine-Dalgarno sequence (SD), and start codons are demarked with dashed boxes. Fusion plasmids were introduced into L. interrogans serovar Lai strain 56601. Transconjugants were incubated in EMJH medium (EMJH) or EMJH with 120 mM sodium chloride (NaCl) for 4 h (B) or in EMJH medium only (C). Fluorescence is shown in arbitrary units. The dashed lines indicate the mean background readings obtained with L. interrogans carrying the empty gfp fusion vector pRAT724. Means ± standard deviations are plotted (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.000; NS, not significant by Welch's t test. S17, 17-bp sequence scrambled.