Figure 1: LIN28 cold shock domain (CSD) and zinc knuckle domain (ZKD) recognize distinct sequence motifs as defined by single-nucleotide-resolution analysis of CLIP data.
Related to Figures S1.
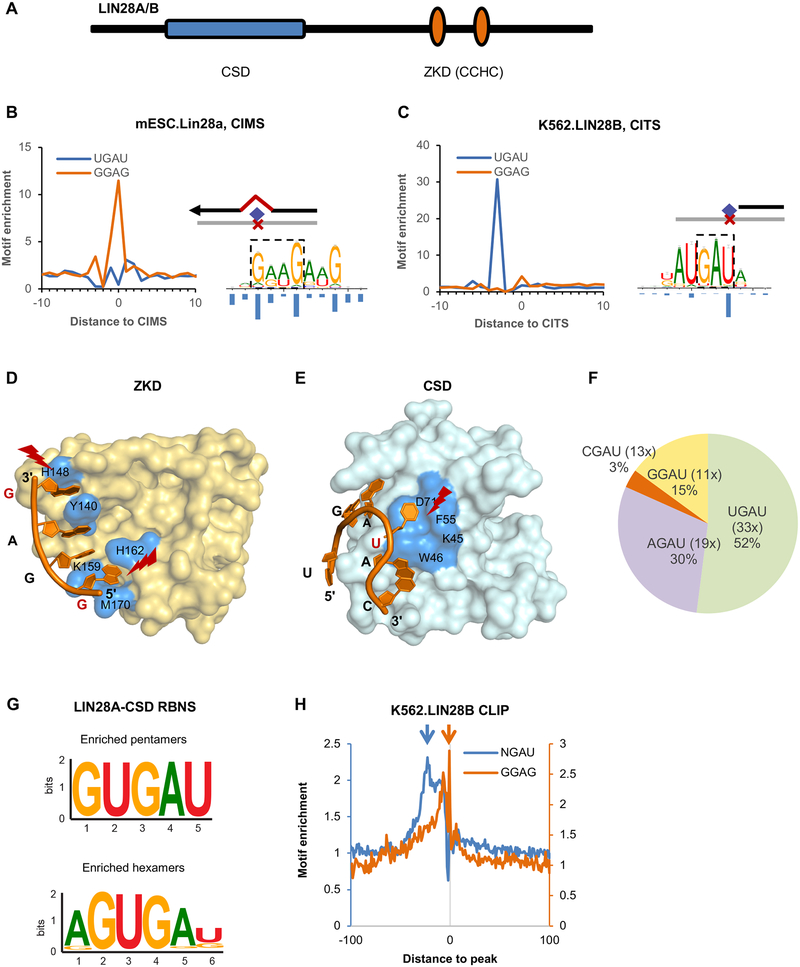
(A) Schematic representation of LIN28 protein domains.
(B, C) The ZKD and CSD binding motifs determined from single-nucleotide-resolution analysis of CLIP data. A GGAG-like motif was identified by modeling sequences around LIN28A CIMS derived from mouse ESCs (B, right panel), and a UGAU motif was determined by modeling sequences around LIN28B CITS derived from K562 cells (C, right panel). The frequency of crosslinking at each motif position is shown under the motif logos. The enrichment of GGAG and UGAU tetramers around CIMS or CITS is shown on the left of each panel.
(D, E) The crystal structure of LIN28A ZKD (D) and CSD (E) in complex with let-7g hairpin (PDB accession: 3TS2). Residues that are in direct contact with RNA are highlighted in blue. The crosslinked nucleotides are indicated in red and highlighted.
(F) Frequency of tetramers conforming to the NGAU consensus in LIN28B eCLIP data from K562 cells. The fold enrichment of each tetramer at the crosslink site in comparison to matched control sequences is shown in the parentheses.
(G) CSD binding motifs identified from RBNS analysis. The most enriched pentamers and hexamers after two rounds of LIN28A CSD selection are shown.
(H) Enrichment of NGAU and GGAG around LIN28 eCLIP tag cluster peaks from K562 cells.