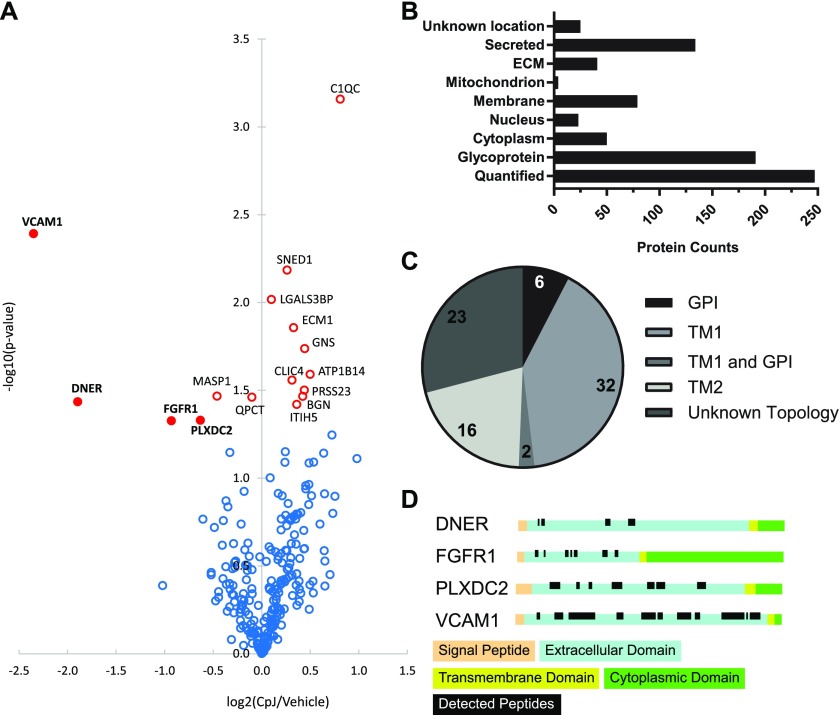
Figure 2. BACE2 secretome in primary glia cultures.
(A) Volcano plot of proteomic analysis of conditioned medium from Bace1−/− cultured glia treated with BACE inhibitor CpJ or vehicle (n = 3). For each relatively quantified protein (representing a dot on the plot), the −log10-transformed t test P-value was plotted against the log2-transformed LFQ intensity ratios of CpJ/vehicle. Proteins with a t test P-value <0.05 are marked with open red circles, whereas proteins with P-value >0.05 are shown in blue. Closed red dots labeled with bold letters denote the four substrate candidates that are reduced by more than 30% in the inhibitor-treated samples. No corrections for multiple hypothesis testing were applied in this discovery experiment. (B) Uniprot subcellular locations of the identified proteins. Glycoproteins were defined according to UniProt Keywords. (C) Topology of membrane proteins according to Uniprot subcellular locations. (D) Uniprot Topology of DNER, FGFR1, PLXDC2, and VCAM1 with mapped identified peptides.