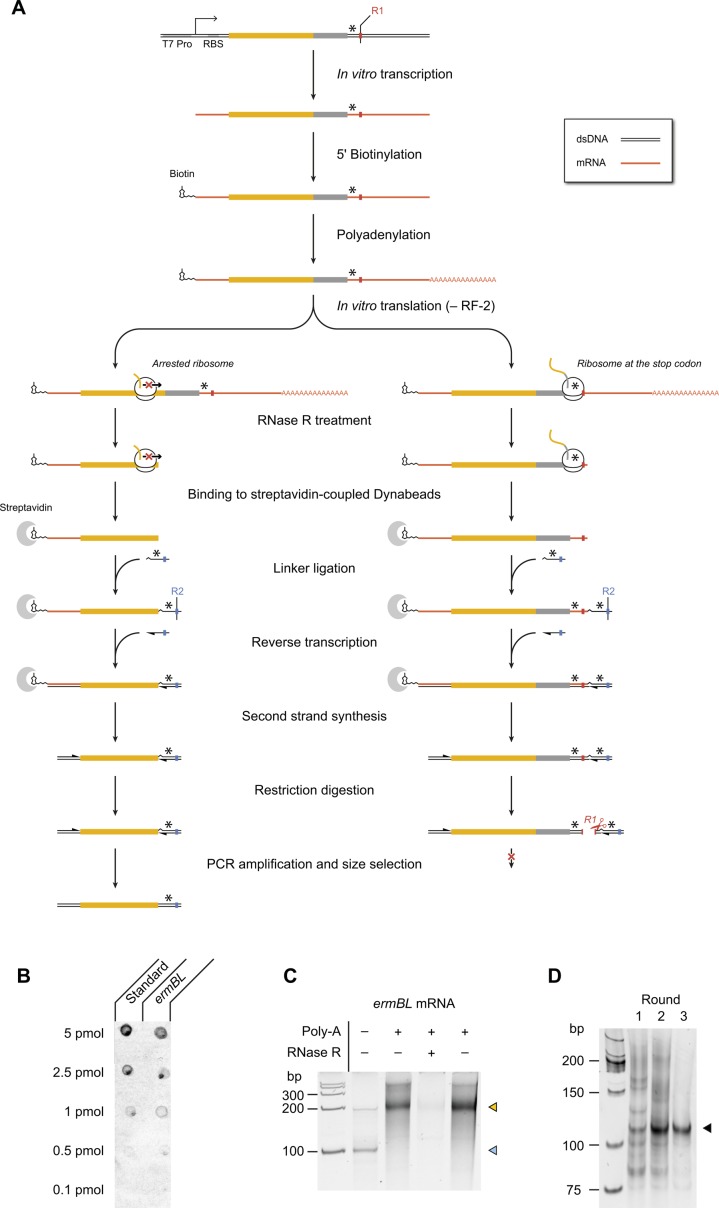
Figure S1. Overview of the inverse toeprinting workflow.
(A) Step-by-step description of the inverse toeprinting methodology. Restriction enzymes used in odd (EcoRV) and even (ApoI) cycles are shown in red and blue, respectively. Stop codons are indicated as asterisks. (B) Efficiency of ermBL mRNA biotinylation measured using a dot blot assay. The standard used is a synthetic biotinylated oligonucleotide, which corresponds to 100% biotinylation efficiency. (C) Efficiency of polyadenylation and digestion by RNase R. The light blue arrow indicates untreated ermBL mRNA and the yellow arrow indicates polyadenylated ermBL mRNA. (D) Amplification of ermBL toeprints following one, two, or three rounds of inverse toeprinting in the presence of Ery.