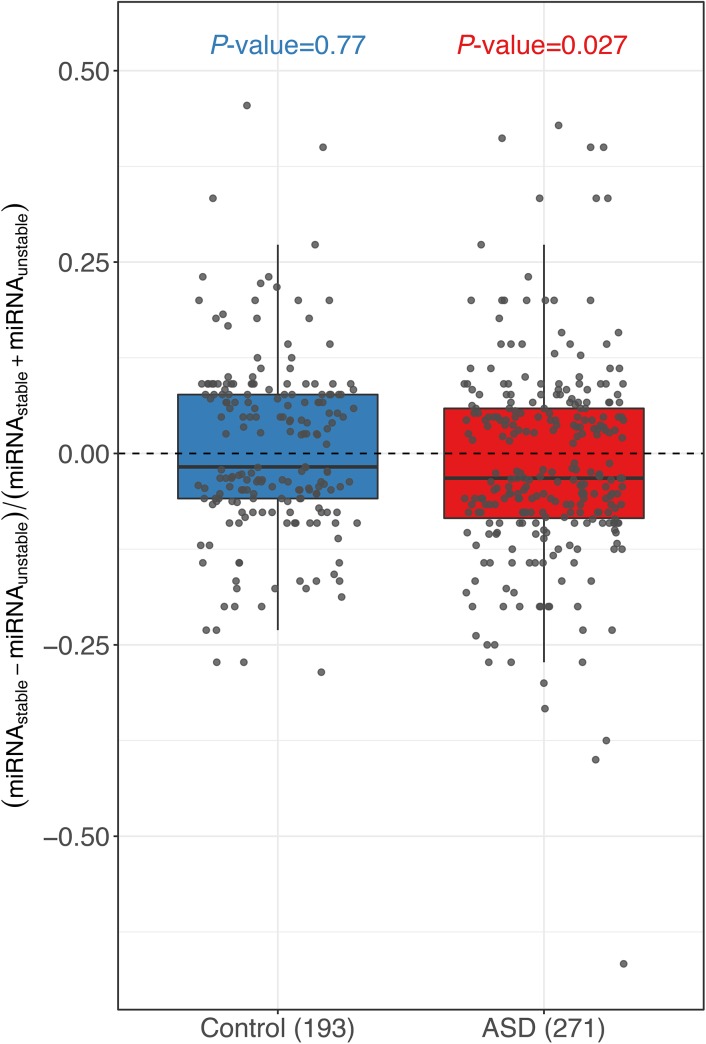
Figure S7. Box plots and scatterplots showing the distribution of miRNA-binding site number difference between the stable and unstable alleles for genes with significant ASD and controls estimated using miRanda.
To validate our findings for miRNA-binding sites using TargetScan (Fig 3B), a similar analysis was performed using a different miRNA-binding site prediction algorithm miRanda (version 3.3a), with parameters miranda mirna.fa target.fa -sc 180 -en 1 -scale 4, in which mirna.fa was downloaded from miRBase (http://www.mirbase.org/). For controls, the difference centered around zero (P-value = 0.77, two-sided Mann–Whitney U test), whereas in ASD genes, unstable alleles tend to possess more miRNA target sites than the stable alleles (P-value = 0.027, two-sided Mann–Whitney U test). Only the genes with ≥10 miRNA-binding sites combining the two alleles together and ≥1 different sites were used.