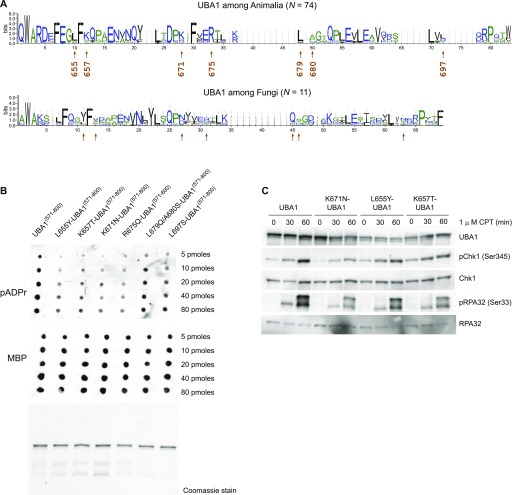
Figure 6. UBA1 residues conserved in animals (but not in fungi) are critical for pADPr binding.
(A) Sequence logos of the alignment of the region shown in Fig 5B. The height of the letters in each position represents the degree of sequence conservation. Colours are used for differentiation among hydrophilic (blue), hydrophobic (black), or neutral amino acids (green). Error bars correspond to the Bayesian 95% confidence interval of the height. Arrows indicate the positions of the amino acids chosen for mutagenesis. These seven amino acids are not conserved between Animalia and Fungi. (B) Increasing amounts (5 to 80 pM) of purified MBPUBA1(571–800) fragments containing the indicated amino acid substitutions were spotted on a nitrocellulose membrane, incubated with purified pADPr chains, and washed. The retention of pADPr was analysed using an anti-pADPr antibody. UBA1 protein fragments used in the pADPr binding assay were revealed using an anti-MBP antibody (middle panel) or resolved by PAGE and stained with Coomassie Brilliant Blue (bottom panel). (C) U2OS cells were transfected with pAIO vector encoding an anti-UBA1 shRNA and an shRNA-resistant UBA1 cDNA encoding wild-type or mutated UBA1, as indicated, and treated with doxycyclin for 3 d. Cells were exposed to 1 μM CPT for increasing amounts of time (as indicated). Indicated proteins were detected by Western blotting.